You are browsing environment: HUMAN GUT
MGYG000004003_02485
Basic Information
help
Species
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides;
CAZyme ID
MGYG000004003_02485
CAZy Family
GT0
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000004003
5653807
MAG
United Kingdom
Europe
Gene Location
Start: 21651;
End: 22595
Strand: +
No EC number prediction in MGYG000004003_02485.
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam13632
Glyco_trans_2_3
9.74e-09
106
282
19
194
Glycosyl transferase family group 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, which are involved in bacterial capsule biosynthesis.
more
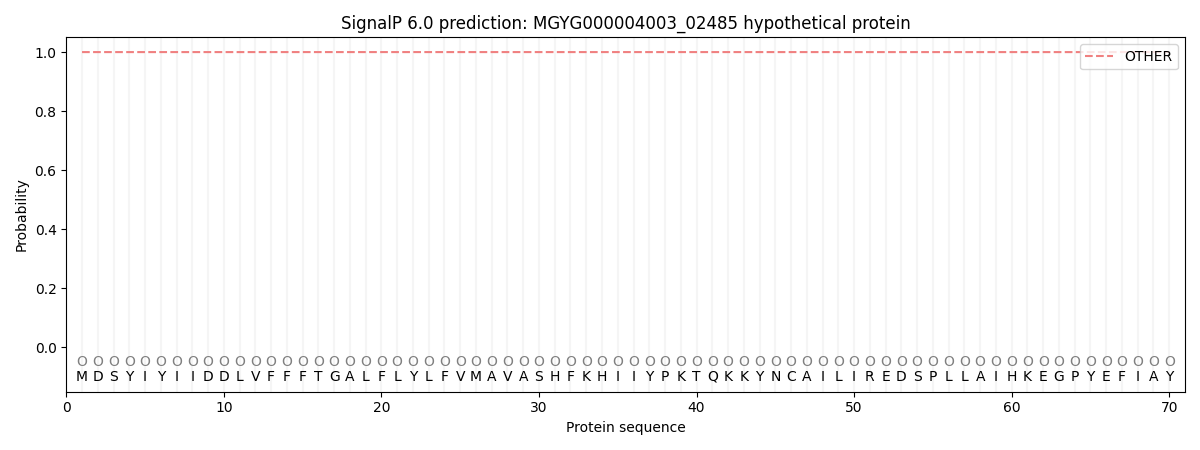
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
1.000052
0.000006
0.000000
0.000000
0.000000
0.000000
start
end
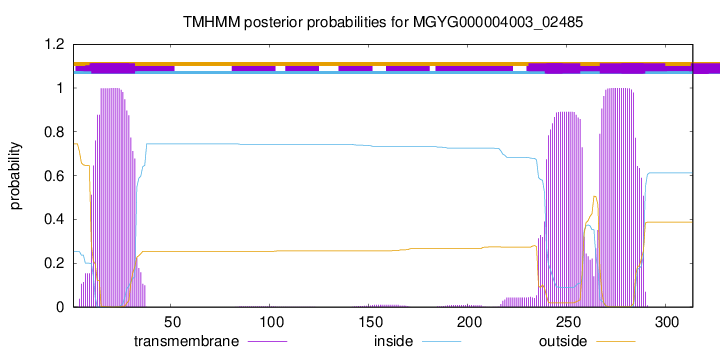
10
32
240
257
267
289