You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004003_02511
You are here: Home > Sequence: MGYG000004003_02511
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
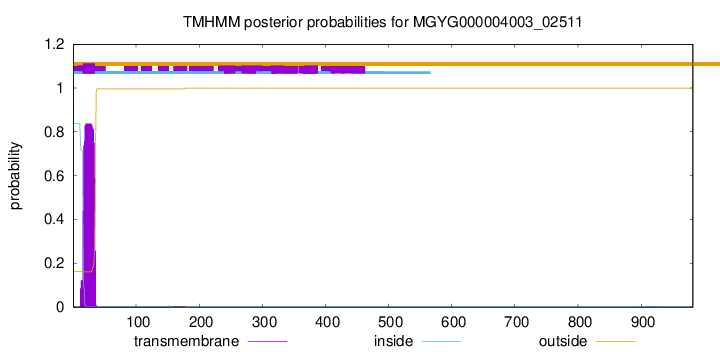
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; | |||||||||||
| CAZyme ID | MGYG000004003_02511 | |||||||||||
| CAZy Family | GH115 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10108; End: 13056 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH115 | 43 | 670 | 4.6e-229 | 0.8421807747489239 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam15979 | Glyco_hydro_115 | 1.11e-171 | 197 | 538 | 1 | 333 | Glycosyl hydrolase family 115. Glyco_hydro_115 is a family of glycoside hydrolases likely to have the activity of xylan a-1,2-glucuronidase, EC:3.2.1.131, or a-(4-O-methyl)-glucuronidase EC:3.2.1.-. |
| pfam17829 | GH115_C | 6.82e-50 | 797 | 977 | 2 | 171 | Gylcosyl hydrolase family 115 C-terminal domain. This domain is found at the C-terminus of glycosyl hydrolase family 115 proteins. This domain has a beta-sandwich fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUR45611.1 | 0.0 | 13 | 982 | 1 | 970 |
| QUR45612.1 | 0.0 | 26 | 980 | 17 | 982 |
| ALJ47615.1 | 0.0 | 5 | 980 | 30 | 1018 |
| QGT72411.1 | 0.0 | 13 | 980 | 1 | 981 |
| EDO12283.1 | 0.0 | 13 | 980 | 1 | 981 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ZMH_A | 3.31e-155 | 54 | 976 | 23 | 930 | Crystalstructure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T [Saccharophagus degradans 2-40],4ZMH_B Crystal structure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T [Saccharophagus degradans 2-40] |
| 5BY3_A | 2.07e-134 | 42 | 649 | 17 | 611 | Anovel family GH115 4-O-Methyl-alpha-glucuronidase, BtGH115A, with specificity for decorated arabinogalactans [Bacteroides thetaiotaomicron VPI-5482] |
| 4C90_A | 1.91e-117 | 11 | 679 | 10 | 662 | Evidencethat GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C90_B Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C91_A Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C91_B Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus] |
| 6NPS_A | 3.36e-110 | 47 | 979 | 12 | 963 | Crystalstructure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus [Amphibacillus xylanus NBRC 15112],6NPS_B Crystal structure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus [Amphibacillus xylanus NBRC 15112] |
| 7PUG_A | 1.18e-104 | 35 | 679 | 7 | 647 | ChainA, xylan alpha-1,2-glucuronidase [uncultured bacterium] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001073 | 0.997774 | 0.000580 | 0.000185 | 0.000183 | 0.000169 |