You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004003_02836
You are here: Home > Sequence: MGYG000004003_02836
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; | |||||||||||
| CAZyme ID | MGYG000004003_02836 | |||||||||||
| CAZy Family | PL38 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 28061; End: 29233 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL38 | 53 | 329 | 7.4e-111 | 0.996415770609319 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05426 | Alginate_lyase | 6.01e-101 | 52 | 329 | 1 | 274 | Alginate lyase. This family contains several bacterial alginate lyase proteins. Alginate is a family of 1-4-linked copolymers of beta -D-mannuronic acid (M) and alpha -L-guluronic acid (G). It is produced by brown algae and by some bacteria belonging to the genera Azotobacter and Pseudomonas. Alginate lyases catalyze the depolymerization of alginates by beta -elimination, generating a molecule containing 4-deoxy-L-erythro-hex-4-enepyranosyluronate at the nonreducing end. This family adopts an all alpha fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIU95437.1 | 3.54e-275 | 8 | 390 | 5 | 387 |
| QNL37221.1 | 7.10e-232 | 1 | 389 | 1 | 388 |
| QUT48713.1 | 2.27e-185 | 29 | 389 | 25 | 385 |
| AFK02440.1 | 1.33e-177 | 6 | 389 | 4 | 386 |
| QRQ56514.1 | 5.06e-177 | 1 | 317 | 1 | 316 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3NFV_A | 4.79e-54 | 39 | 332 | 34 | 331 | Crystalstructure of an alginate lyase (BACOVA_01668) from Bacteroides ovatus at 1.95 A resolution [Bacteroides ovatus ATCC 8483],3NNB_A Crystal structure of an alginate lyase (BACOVA_01668) from Bacteroides ovatus at 1.60 A resolution [Bacteroides ovatus ATCC 8483] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
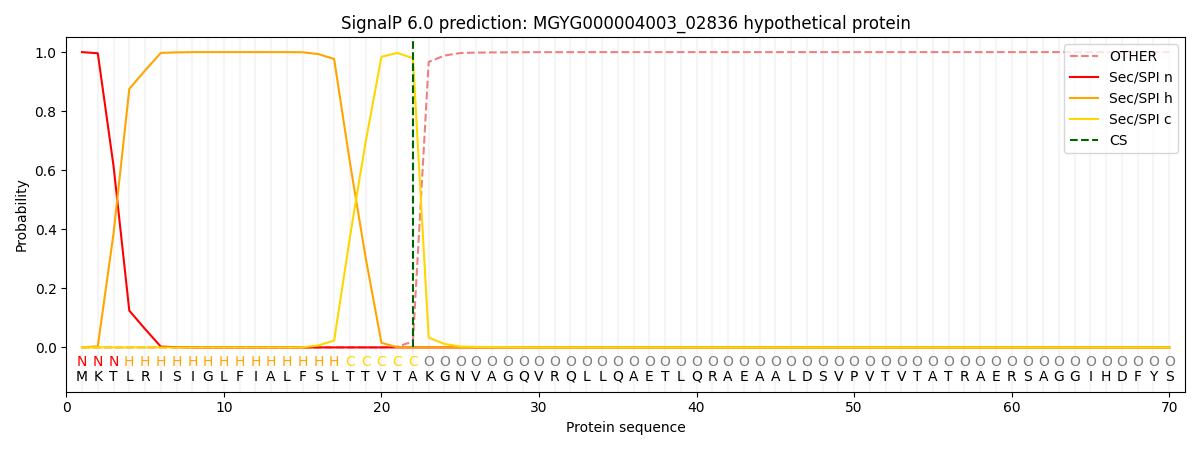
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000288 | 0.999031 | 0.000201 | 0.000154 | 0.000150 | 0.000146 |
