You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004021_00688
You are here: Home > Sequence: MGYG000004021_00688
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA11471 sp900542765 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA11471; UBA11471; UBA11471 sp900542765 | |||||||||||
| CAZyme ID | MGYG000004021_00688 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 157689; End: 159257 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 85 | 471 | 1.5e-84 | 0.9230769230769231 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 1.15e-77 | 11 | 514 | 31 | 523 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 7.22e-20 | 192 | 502 | 59 | 316 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PLN03003 | PLN03003 | 2.12e-14 | 197 | 384 | 102 | 276 | Probable polygalacturonase At3g15720 |
| PLN03010 | PLN03010 | 9.29e-10 | 243 | 384 | 162 | 295 | polygalacturonase |
| PLN02218 | PLN02218 | 9.73e-09 | 60 | 384 | 68 | 330 | polygalacturonase ADPG |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDH54501.1 | 7.84e-240 | 22 | 521 | 20 | 522 |
| QNL41099.1 | 1.11e-239 | 39 | 521 | 37 | 522 |
| SCV08391.1 | 1.58e-239 | 39 | 521 | 37 | 522 |
| QRQ55782.1 | 1.58e-239 | 39 | 521 | 37 | 522 |
| ALJ48973.1 | 1.58e-239 | 39 | 521 | 37 | 522 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3JUR_A | 1.37e-82 | 50 | 486 | 18 | 433 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
| 5OLP_A | 2.17e-68 | 44 | 482 | 29 | 441 | Galacturonidase[Bacteroides thetaiotaomicron VPI-5482],5OLP_B Galacturonidase [Bacteroides thetaiotaomicron VPI-5482] |
| 2UVE_A | 5.17e-34 | 60 | 421 | 157 | 512 | Structureof Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVE_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVF_A Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica],2UVF_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica] |
| 1BHE_A | 7.67e-17 | 76 | 433 | 26 | 333 | ChainA, POLYGALACTURONASE [Pectobacterium carotovorum] |
| 4MXN_A | 1.86e-12 | 57 | 306 | 21 | 214 | Crystalstructure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_B Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_C Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_D Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7PZL3 | 9.66e-42 | 31 | 467 | 39 | 424 | Probable polygalacturonase OS=Vitis vinifera OX=29760 GN=GSVIVT00026920001 PE=1 SV=1 |
| P27644 | 7.27e-35 | 216 | 456 | 3 | 249 | Polygalacturonase OS=Rhizobium radiobacter OX=358 GN=pgl PE=2 SV=1 |
| P15922 | 6.74e-33 | 60 | 421 | 152 | 505 | Exo-poly-alpha-D-galacturonosidase OS=Dickeya chrysanthemi OX=556 GN=pehX PE=1 SV=1 |
| P58598 | 4.13e-18 | 82 | 510 | 90 | 507 | Polygalacturonase OS=Ralstonia solanacearum (strain GMI1000) OX=267608 GN=pglA PE=3 SV=1 |
| Q9LW07 | 1.25e-17 | 197 | 432 | 102 | 326 | Probable polygalacturonase At3g15720 OS=Arabidopsis thaliana OX=3702 GN=At3g15720 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
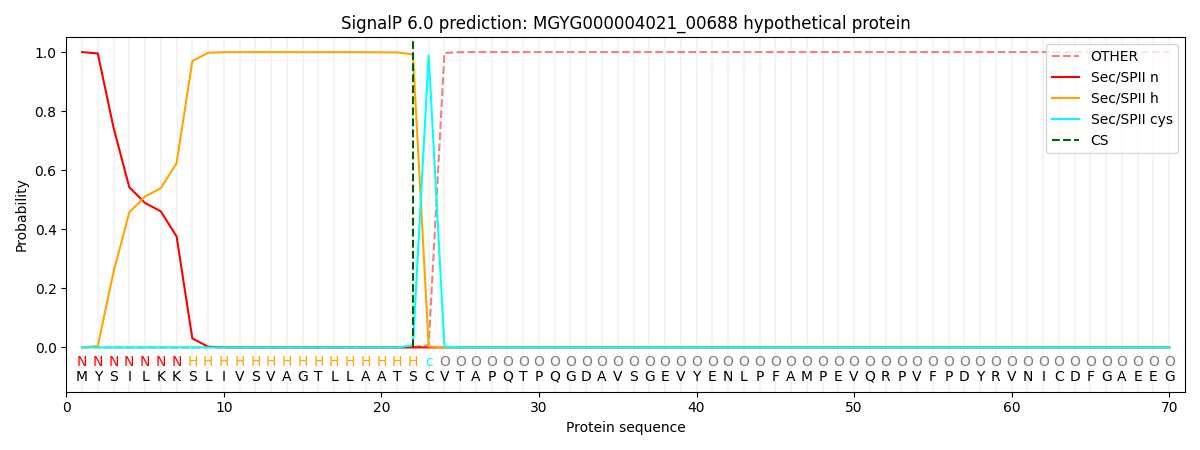
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000013 | 1.000030 | 0.000000 | 0.000000 | 0.000000 |
