You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004037_00492
You are here: Home > Sequence: MGYG000004037_00492
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
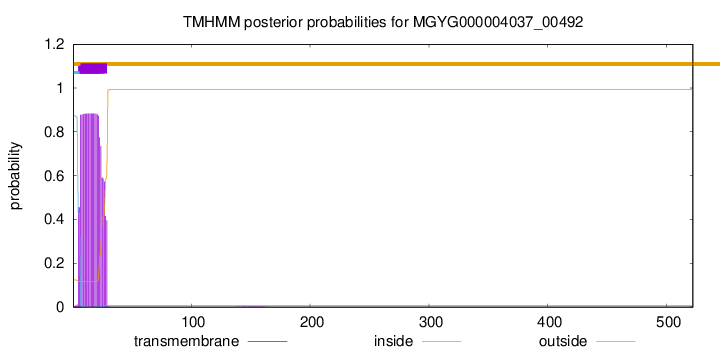
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA11471; UBA11471; | |||||||||||
| CAZyme ID | MGYG000004037_00492 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 85284; End: 86852 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 223 | 491 | 2.4e-45 | 0.9131944444444444 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 7.11e-17 | 223 | 476 | 86 | 371 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02773 | PLN02773 | 4.79e-16 | 219 | 498 | 4 | 283 | pectinesterase |
| PLN02488 | PLN02488 | 2.07e-14 | 215 | 478 | 190 | 459 | probable pectinesterase/pectinesterase inhibitor |
| PLN02197 | PLN02197 | 2.58e-12 | 219 | 478 | 274 | 540 | pectinesterase |
| PLN02176 | PLN02176 | 5.92e-12 | 222 | 498 | 41 | 314 | putative pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJD94164.1 | 1.56e-165 | 36 | 520 | 19 | 491 |
| QCD38260.1 | 6.05e-159 | 48 | 521 | 31 | 497 |
| QCP71948.1 | 6.05e-159 | 48 | 521 | 31 | 497 |
| QUT59621.1 | 5.68e-156 | 40 | 520 | 21 | 498 |
| QQY40500.1 | 5.68e-156 | 40 | 520 | 21 | 498 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
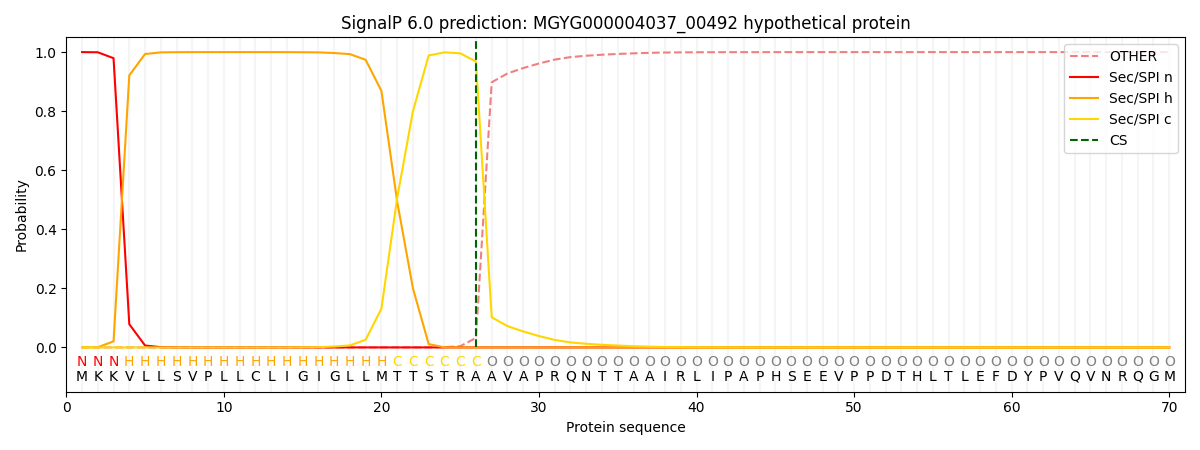
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000416 | 0.998702 | 0.000283 | 0.000212 | 0.000190 | 0.000180 |