You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004037_01032
You are here: Home > Sequence: MGYG000004037_01032
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA11471; UBA11471; | |||||||||||
| CAZyme ID | MGYG000004037_01032 | |||||||||||
| CAZy Family | PL11 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12094; End: 14880 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL11 | 287 | 866 | 1.1e-232 | 0.9777015437392796 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd10318 | RGL11 | 0.0 | 287 | 859 | 2 | 564 | Rhamnogalacturonan lyase of the polysaccharide lyase family 11. The rhamnogalacturonan lyase of the polysaccharide lyase family 11 (RGL11) cleaves glycoside bonds in polygalacturonan as well as RG (rhamnogalacturonan) type-I through a beta-elimination reaction. Functionally characterized members of this family, YesW and YesX from Bacillus subtilis, cleave glycoside bonds between rhamnose and galacturonic acid residues in the RG-I region of plant cell wall pectin. YesW and YesX work synergistically, with YesW cleaving the glycoside bond of the RG chain endolytically, and YesX converting the resultant oligosaccharides through an exotype reaction. This domain is sometimes found in architectures with non-catalytic carbohydrate-binding modules (CBMs). There are two types of RG lyases, which both cleave the alpha-1,4 bonds of the RG-I main chain through a beta-elimination reaction, but belong to two structurally unrelated polysaccharide lyase (PL) families, 4 and 11. |
| pfam18370 | RGI_lyase | 2.48e-27 | 287 | 367 | 4 | 82 | Rhamnogalacturonan I lyases beta-sheet domain. This is the beta-sheet domain found in rhamnogalacturonan (RG) lyases, which are responsible for an initial cleavage of the RG type I (RG-I) region of plant cell wall pectin. Polysaccharide lyase family 11 carrying this domain, such as YesW (EC:4.2.2.23) and YesX (EC:4.2.2.24), cleave glycoside bonds between rhamnose and galacturonic acid residues in RG-I through a beta-elimination reaction. Other family members carrying this domain are hemagglutinin A, lysine gingipain (Kgp) and Chitinase C (EC:3.2.1.14). |
| pfam13290 | CHB_HEX_C_1 | 6.39e-06 | 210 | 266 | 12 | 67 | Chitobiase/beta-hexosaminidase C-terminal domain. |
| pfam13287 | Fn3_assoc | 2.23e-04 | 210 | 264 | 1 | 54 | Fn3 associated. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT74217.1 | 3.43e-295 | 197 | 923 | 414 | 1132 |
| QUT73808.1 | 2.81e-224 | 282 | 864 | 21 | 619 |
| QCD43243.1 | 2.29e-207 | 284 | 884 | 17 | 624 |
| AEE95353.1 | 3.70e-203 | 287 | 868 | 41 | 610 |
| SCV07134.1 | 1.52e-198 | 287 | 868 | 30 | 627 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4CAG_A | 1.35e-190 | 287 | 867 | 12 | 581 | Bacilluslicheniformis Rhamnogalacturonan Lyase PL11 [Bacillus licheniformis] |
| 2Z8R_A | 5.08e-189 | 287 | 863 | 6 | 570 | Crystalstructure of rhamnogalacturonan lyase YesW at 1.40 A resolution [Bacillus subtilis],2Z8R_B Crystal structure of rhamnogalacturonan lyase YesW at 1.40 A resolution [Bacillus subtilis],2Z8S_A Crystal structure of rhamnogalacturonan lyase YesW complexed with digalacturonic acid [Bacillus subtilis],2Z8S_B Crystal structure of rhamnogalacturonan lyase YesW complexed with digalacturonic acid [Bacillus subtilis],2ZUX_A Crystal structure of rhamnogalacturonan lyase YesW complexed with rhamnose [Bacillus subtilis],2ZUX_B Crystal structure of rhamnogalacturonan lyase YesW complexed with rhamnose [Bacillus subtilis] |
| 2ZUY_A | 2.96e-181 | 287 | 861 | 9 | 589 | Crystalstructure of exotype rhamnogalacturonan lyase YesX [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31526 | 7.37e-188 | 287 | 863 | 43 | 607 | Rhamnogalacturonan endolyase YesW OS=Bacillus subtilis (strain 168) OX=224308 GN=yesW PE=1 SV=1 |
| O31527 | 1.24e-180 | 287 | 861 | 9 | 589 | Rhamnogalacturonan exolyase YesX OS=Bacillus subtilis (strain 168) OX=224308 GN=yesX PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
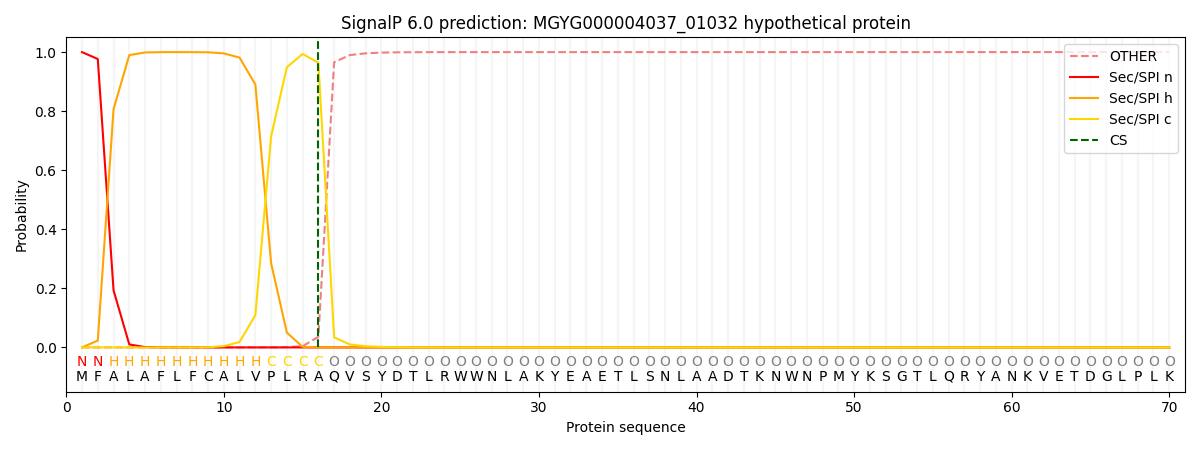
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000823 | 0.998446 | 0.000205 | 0.000165 | 0.000153 | 0.000157 |
