You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004037_01780
You are here: Home > Sequence: MGYG000004037_01780
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
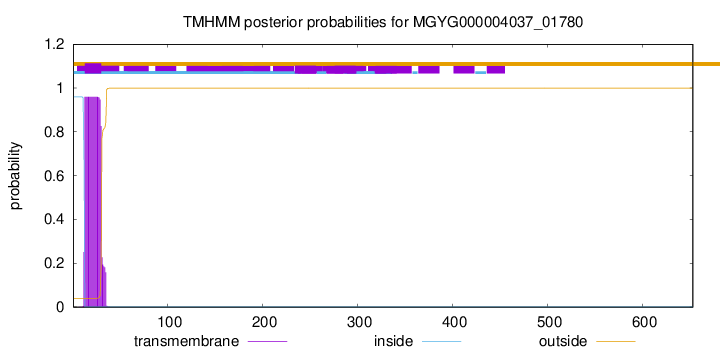
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA11471; UBA11471; | |||||||||||
| CAZyme ID | MGYG000004037_01780 | |||||||||||
| CAZy Family | CBM62 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1774; End: 3735 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01841 | Transglut_core | 0.008 | 231 | 287 | 48 | 108 | Transglutaminase-like superfamily. This family includes animal transglutaminases and other bacterial proteins of unknown function. Sequence conservation in this superfamily primarily involves three motifs that centre around conserved cysteine, histidine, and aspartate residues that form the catalytic triad in the structurally characterized transglutaminase, the human blood clotting factor XIIIa'. On the basis of the experimentally demonstrated activity of the Methanobacterium phage pseudomurein endoisopeptidase, it is proposed that many, if not all, microbial homologs of the transglutaminases are proteases and that the eukaryotic transglutaminases have evolved from an ancestral protease. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ49056.1 | 1.45e-268 | 38 | 653 | 26 | 638 |
| QRQ55862.1 | 1.67e-268 | 38 | 653 | 30 | 642 |
| SCV08471.1 | 1.67e-268 | 38 | 653 | 30 | 642 |
| ANU56555.1 | 2.92e-268 | 38 | 653 | 26 | 638 |
| QQR18597.1 | 2.92e-268 | 38 | 653 | 26 | 638 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
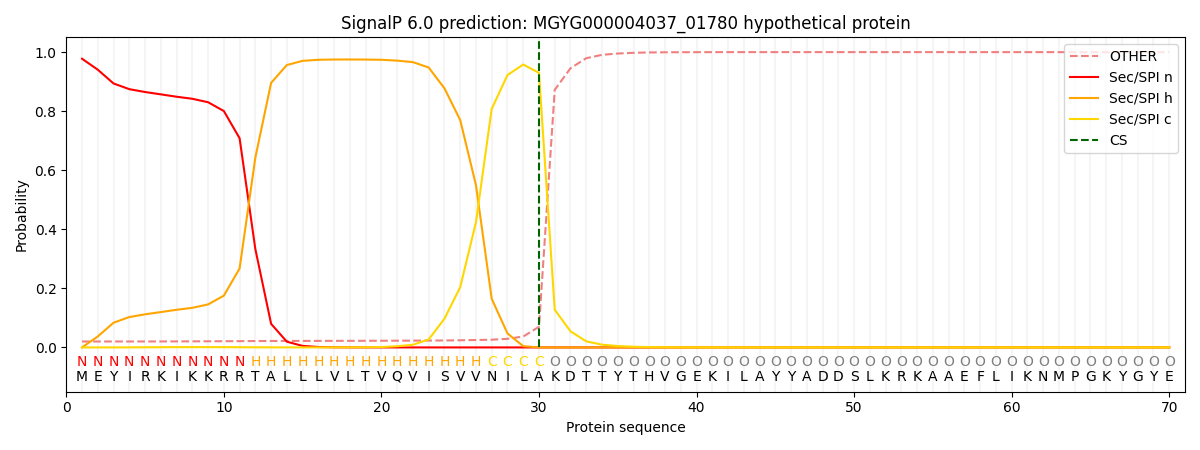
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.023619 | 0.972491 | 0.002959 | 0.000342 | 0.000265 | 0.000288 |