You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004043_00211
You are here: Home > Sequence: MGYG000004043_00211
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; UBA7173; | |||||||||||
| CAZyme ID | MGYG000004043_00211 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 93401; End: 95953 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 544 | 750 | 2.9e-20 | 0.7466216216216216 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00704 | Glyco_hydro_18 | 9.62e-15 | 541 | 744 | 68 | 307 | Glycosyl hydrolases family 18. |
| pfam13385 | Laminin_G_3 | 2.02e-13 | 51 | 165 | 19 | 148 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| smart00636 | Glyco_18 | 1.26e-12 | 493 | 744 | 36 | 334 | Glyco_18 domain. |
| cd06548 | GH18_chitinase | 2.95e-08 | 541 | 744 | 89 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| COG3325 | ChiA | 2.46e-05 | 540 | 745 | 130 | 424 | Chitinase, GH18 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFQ12875.1 | 7.42e-138 | 25 | 791 | 26 | 779 |
| QUT75436.1 | 1.48e-125 | 26 | 797 | 25 | 781 |
| QDO70362.1 | 1.18e-93 | 25 | 791 | 22 | 781 |
| AHF11971.1 | 3.32e-88 | 3 | 782 | 2 | 766 |
| BCI61676.1 | 1.77e-78 | 3 | 791 | 2 | 773 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
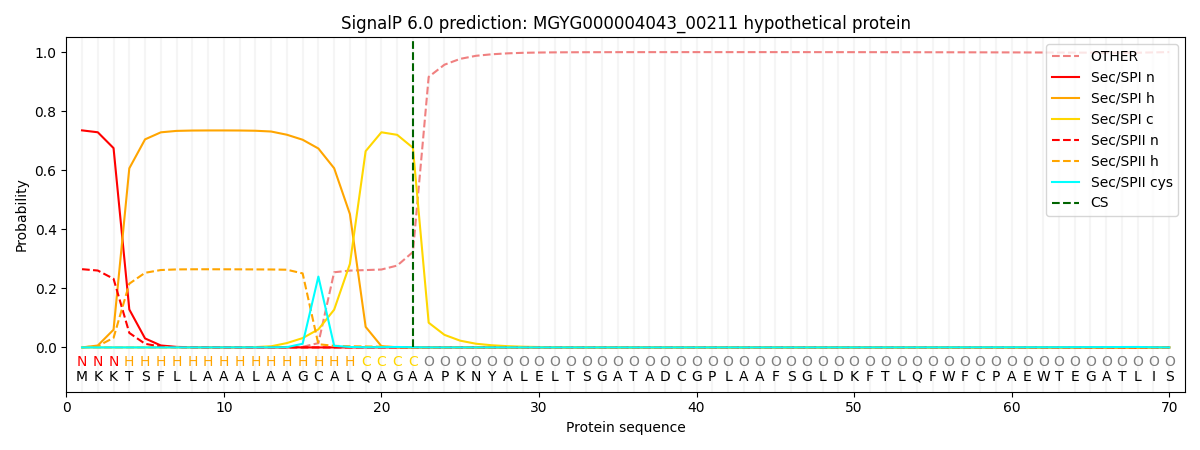
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000555 | 0.725260 | 0.273204 | 0.000426 | 0.000295 | 0.000232 |
