You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004043_00311
You are here: Home > Sequence: MGYG000004043_00311
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; UBA7173; | |||||||||||
| CAZyme ID | MGYG000004043_00311 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 105444; End: 109649 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02432 | PLN02432 | 1.06e-17 | 1076 | 1252 | 120 | 292 | putative pectinesterase |
| COG4677 | PemB | 6.79e-15 | 937 | 1168 | 73 | 332 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02773 | PLN02773 | 7.88e-14 | 1083 | 1252 | 136 | 303 | pectinesterase |
| pfam01095 | Pectinesterase | 8.86e-14 | 962 | 1248 | 15 | 298 | Pectinesterase. |
| PLN02170 | PLN02170 | 6.32e-12 | 968 | 1252 | 243 | 517 | probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASB37302.1 | 5.83e-248 | 392 | 1397 | 173 | 1156 |
| ANU64595.1 | 5.83e-248 | 392 | 1397 | 173 | 1156 |
| QQR08039.1 | 5.83e-248 | 392 | 1397 | 173 | 1156 |
| AWK06216.1 | 2.06e-56 | 844 | 1339 | 494 | 976 |
| QUT74167.1 | 3.29e-56 | 768 | 1400 | 819 | 1434 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2NSP_A | 3.07e-16 | 957 | 1254 | 11 | 340 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 5C1E_A | 1.01e-15 | 992 | 1213 | 29 | 270 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
| 3UW0_A | 3.02e-15 | 982 | 1192 | 62 | 306 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
| 5C1C_A | 4.47e-15 | 992 | 1213 | 29 | 270 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
| 4PMH_A | 1.21e-08 | 1083 | 1169 | 192 | 294 | Thestructure of rice weevil pectin methyl esterase [Sitophilus oryzae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A2QK82 | 4.51e-15 | 992 | 1213 | 57 | 298 | Probable pectinesterase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pmeA PE=3 SV=1 |
| Q5B7U0 | 1.50e-13 | 992 | 1252 | 55 | 321 | Pectinesterase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=pmeA PE=1 SV=1 |
| Q9ZQA4 | 2.20e-13 | 989 | 1252 | 60 | 333 | Putative pectinesterase 14 OS=Arabidopsis thaliana OX=3702 GN=PME14 PE=2 SV=1 |
| Q9LUL8 | 2.81e-12 | 977 | 1252 | 685 | 958 | Putative pectinesterase/pectinesterase inhibitor 26 OS=Arabidopsis thaliana OX=3702 GN=PME26 PE=2 SV=1 |
| O04887 | 6.38e-12 | 958 | 1252 | 212 | 499 | Pectinesterase 2 OS=Citrus sinensis OX=2711 GN=PECS-2.1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
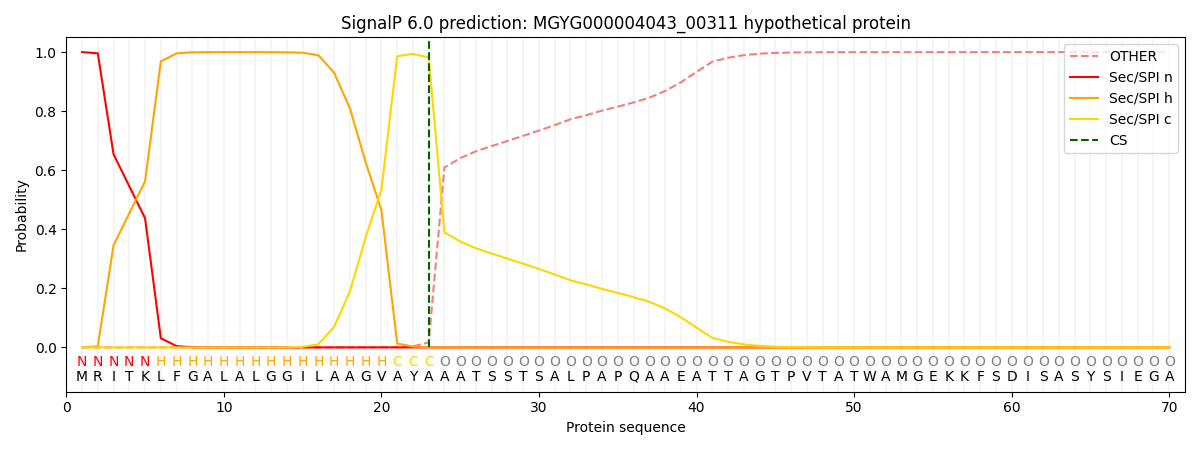
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000375 | 0.998772 | 0.000206 | 0.000246 | 0.000198 | 0.000173 |

