You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004044_02002
You are here: Home > Sequence: MGYG000004044_02002
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS1633 sp900553645 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-74; UMGS1633; UMGS1633 sp900553645 | |||||||||||
| CAZyme ID | MGYG000004044_02002 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2324; End: 5002 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 4 | 461 | 9e-48 | 0.5106382978723404 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 7.84e-13 | 62 | 538 | 77 | 515 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 4.86e-08 | 64 | 425 | 61 | 443 | beta-D-glucuronidase; Provisional |
| pfam00703 | Glyco_hydro_2 | 3.48e-05 | 182 | 276 | 2 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam02836 | Glyco_hydro_2_C | 2.16e-04 | 282 | 425 | 5 | 156 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK10340 | ebgA | 3.42e-04 | 244 | 433 | 281 | 480 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQR31292.1 | 4.00e-286 | 4 | 863 | 2 | 865 |
| QOV18688.1 | 9.06e-286 | 6 | 892 | 4 | 905 |
| ASB42024.1 | 3.45e-282 | 23 | 863 | 1 | 845 |
| ANU54745.1 | 3.45e-282 | 23 | 863 | 1 | 845 |
| QTE72117.1 | 1.13e-272 | 2 | 882 | 1 | 880 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6D89_A | 3.00e-09 | 3 | 426 | 35 | 439 | Bacteroidesuniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_B Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_C Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_D Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis] |
| 6D1N_A | 5.26e-09 | 56 | 426 | 93 | 447 | Apostructure of Bacteroides uniformis Beta-glucuronidase 1 [Bacteroides uniformis],6D1N_B Apo structure of Bacteroides uniformis Beta-glucuronidase 1 [Bacteroides uniformis],6D41_A Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone [Bacteroides uniformis],6D41_B Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone [Bacteroides uniformis],6D6W_A Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_B Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_C Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_D Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D7F_A Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_B Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_C Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_D Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_E Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_F Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
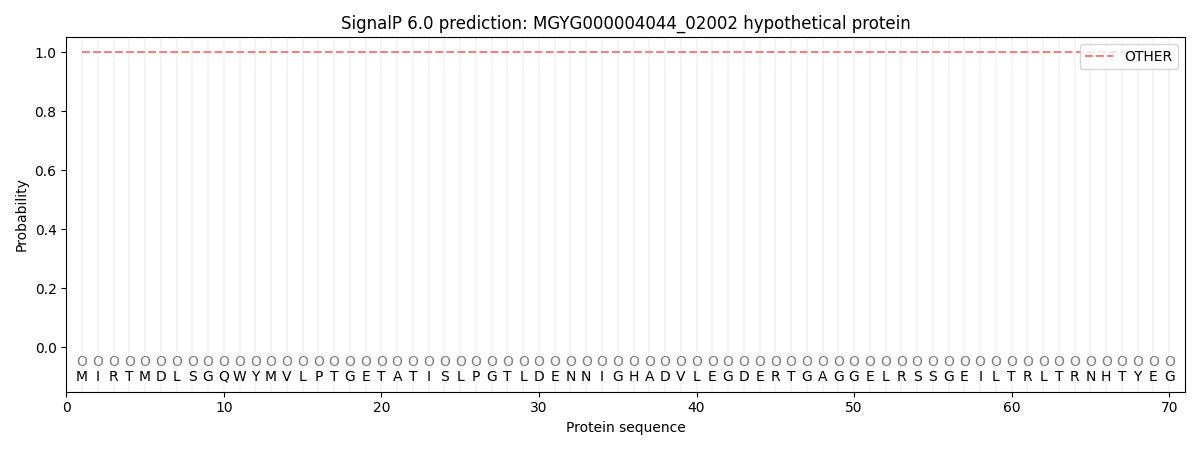
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000058 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
