You are browsing environment: HUMAN GUT
MGYG000004053_00486
Basic Information
help
Species
Lineage
Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312;
CAZyme ID
MGYG000004053_00486
CAZy Family
GH39
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
481
53820.55
9.0283
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000004053
2223892
MAG
United Kingdom
Europe
Gene Location
Start: 22847;
End: 24292
Strand: -
No EC number prediction in MGYG000004053_00486.
Family
Start
End
Evalue
family coverage
GH39
93
260
9.6e-26
0.41067285382830626
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam01229
Glyco_hydro_39
7.10e-09
57
267
21
263
Glycosyl hydrolases family 39.
more
pfam02449
Glyco_hydro_42
3.25e-06
91
176
34
138
Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues.
more
smart00633
Glyco_10
4.66e-04
91
206
3
124
Glycosyl hydrolase family 10.
more
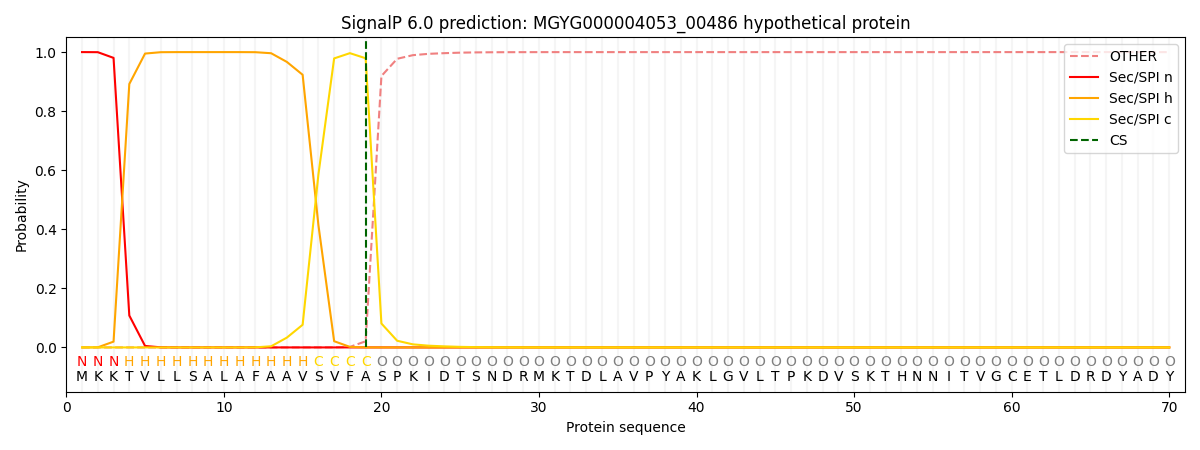
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000286
0.999056
0.000187
0.000159
0.000151
0.000141
There is no transmembrane helices in MGYG000004053_00486.