You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004053_00497
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312;
|
| CAZyme ID |
MGYG000004053_00497
|
| CAZy Family |
GH110 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 788 |
|
88829.99 |
9.4844 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000004053 |
2223892 |
MAG |
United Kingdom |
Europe |
|
| Gene Location |
Start: 1864;
End: 4230
Strand: +
|
No EC number prediction in MGYG000004053_00497.
| Family |
Start |
End |
Evalue |
family coverage |
| GH110 |
225 |
757 |
2.2e-49 |
0.9908759124087592 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam13229
|
Beta_helix |
8.08e-07 |
590 |
758 |
7 |
157 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229
|
Beta_helix |
0.005 |
587 |
641 |
96 |
152 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
Q64MU6
|
6.82e-21 |
222 |
726 |
26 |
548 |
Alpha-1,3-galactosidase A OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=glaA PE=3 SV=1 |
|
B2UL12
|
1.52e-09 |
396 |
734 |
257 |
586 |
Alpha-1,3-galactosidase A OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=glaA PE=3 SV=1 |
This protein is predicted as LIPO
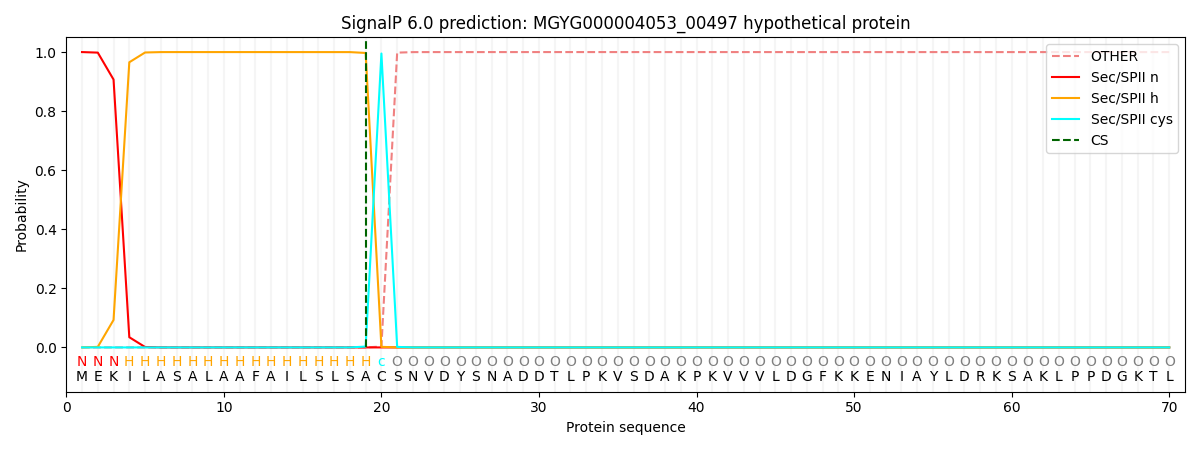
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000000
|
0.000000
|
1.000064
|
0.000000
|
0.000000
|
0.000000
|
There is no transmembrane helices in MGYG000004053_00497.

