You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004055_00468
You are here: Home > Sequence: MGYG000004055_00468
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Eubacterium_G sp900548465 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eubacterium_G; Eubacterium_G sp900548465 | |||||||||||
| CAZyme ID | MGYG000004055_00468 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 100390; End: 103164 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 98 | 382 | 2.8e-90 | 0.9961977186311787 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.59e-25 | 117 | 381 | 21 | 268 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 6.25e-18 | 117 | 429 | 71 | 368 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| cd04084 | CBM6_xylanase-like | 4.85e-17 | 510 | 640 | 1 | 123 | Carbohydrate Binding Module 6 (CBM6); many are appended to glycoside hydrolase (GH) family 11 and GH43 xylanase domains. This family includes carbohydrate binding module 6 (CBM6) domains that are appended mainly to glycoside hydrolase (GH) family domains, including GH3, GH11, and GH43 domains. These CBM6s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. Examples of proteins having CMB6s belonging to this family are Microbispora bispora GghA, a 1,4-beta-D-glucan glucohydrolase (GH3); Clostridium thermocellum xylanase U (GH11), and Penicillium purpurogenum ABF3, a bifunctional alpha-L-arabinofuranosidase/xylobiohydrolase (GH43). GH3 comprises enzymes with activities including beta-glucosidase (hydrolyzes beta-galactosidase) and beta-xylosidase (hydrolyzes 1,4-beta-D-xylosidase). GH11 family comprises enzymes with xylanase (endo-1,4-beta-xylanase) activity which catalyze the hydrolysis of beta-1,4 bonds of xylan, the major component of hemicelluloses, to generate xylooligosaccharides and xylose. GH43 includes beta-xylosidases and beta-xylanases, using aryl-glycosides as substrates. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. |
| smart00606 | CBD_IV | 3.59e-13 | 512 | 640 | 8 | 129 | Cellulose Binding Domain Type IV. |
| pfam03422 | CBM_6 | 1.66e-10 | 514 | 642 | 2 | 125 | Carbohydrate binding module (family 6). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCJ97694.1 | 1.60e-136 | 41 | 443 | 46 | 435 |
| BCK01129.1 | 1.96e-106 | 41 | 577 | 100 | 616 |
| VEI36204.1 | 4.62e-95 | 41 | 454 | 44 | 420 |
| AZS15177.1 | 2.05e-88 | 41 | 448 | 47 | 419 |
| QQY27184.1 | 4.54e-81 | 19 | 481 | 825 | 1250 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1EQP_A | 5.26e-12 | 53 | 329 | 5 | 261 | Exo-b-(1,3)-glucanaseFrom Candida Albicans [Candida albicans] |
| 3N9K_A | 7.19e-12 | 53 | 329 | 10 | 266 | F229A/E292SDouble Mutant of Exo-beta-1,3-glucanase from Candida albicans in Complex with Laminaritriose at 1.7 A [Candida albicans] |
| 1CZ1_A | 9.29e-12 | 53 | 329 | 5 | 261 | Exo-b-(1,3)-glucanaseFrom Candida Albicans At 1.85 A Resolution [Candida albicans],1EQC_A Exo-b-(1,3)-glucanase From Candida Albicans In Complex With Castanospermine At 1.85 A [Candida albicans] |
| 3O6A_A | 9.55e-12 | 53 | 329 | 10 | 266 | F144Y/F258YDouble Mutant of Exo-beta-1,3-glucanase from Candida albicans at 2 A [Candida albicans] |
| 4M80_A | 9.55e-12 | 53 | 329 | 10 | 266 | Thestructure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans at 1.85A resolution [Candida albicans SC5314],4M81_A The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans complexed with 1-fluoro-alpha-D-glucopyranoside (donor) and p-nitrophenyl beta-D-glucopyranoside (acceptor) at 1.86A resolution [Candida albicans SC5314],4M82_A The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans complexed with p-nitrophenyl-gentiobioside (product) at 1.6A resolution [Candida albicans SC5314] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| W8QRE4 | 3.22e-23 | 42 | 317 | 3 | 248 | Beta-xylosidase OS=Phanerodontia chrysosporium OX=2822231 GN=Xyl5 PE=1 SV=2 |
| Q8NKF9 | 6.72e-15 | 59 | 317 | 37 | 281 | Glucan 1,3-beta-glucosidase OS=Candida oleophila OX=45573 GN=EXG1 PE=3 SV=1 |
| O93983 | 9.06e-15 | 59 | 329 | 42 | 291 | Glucan 1,3-beta-glucosidase 2 OS=Wickerhamomyces anomalus OX=4927 GN=EXG2 PE=3 SV=1 |
| Q7Z9L3 | 1.38e-14 | 59 | 332 | 31 | 279 | Glucan 1,3-beta-glucosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=exgA PE=1 SV=1 |
| B8N151 | 1.38e-14 | 59 | 332 | 31 | 279 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=exgA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
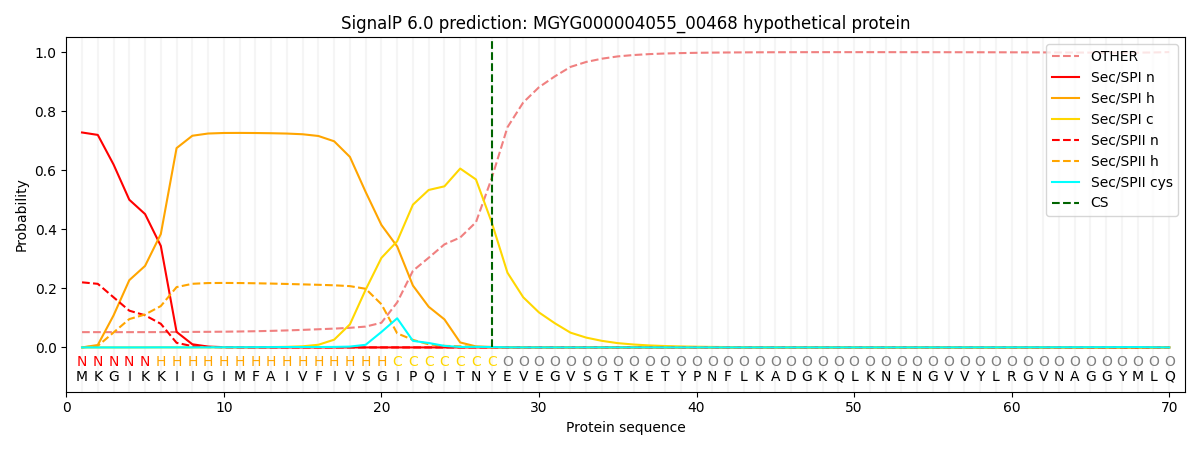
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.056895 | 0.715696 | 0.226315 | 0.000360 | 0.000322 | 0.000393 |

