You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004056_01120
You are here: Home > Sequence: MGYG000004056_01120
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes_A ihumii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes_A; Alistipes_A ihumii | |||||||||||
| CAZyme ID | MGYG000004056_01120 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9268; End: 10698 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 106 | 404 | 4.6e-38 | 0.728538283062645 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01229 | Glyco_hydro_39 | 3.42e-18 | 128 | 343 | 106 | 307 | Glycosyl hydrolases family 39. |
| cd21510 | agarase_cat | 1.40e-06 | 182 | 259 | 100 | 187 | alpha-beta barrel catalytic domain of agarase, such as GH86-like endo-acting agarases identified in non-marine organisms. Typically, agarases (E.C. 3.2.1.81) are found in ocean-dwelling bacteria since agarose is a principle component of red algae cell wall polysaccharides. Agarose is a linear polymer of alternating D-galactose and 3,6-anhydro-L-galactopyranose. Endo-acting agarases, such as glycoside hydrolase 16 (GH16) and GH86 hydrolyze internal beta-1,4 linkages. GH86-like endo-acting agarase of this protein family has been identified in the human intestinal bacterium Bacteroides uniformis. This acquired metabolic pathway, as demonstrated by the prevalence of agar-specific genetic cluster called polysaccharide utilization loci (PULs), varies considerably between human populations, being much more prevalent in a Japanese sample than in North America, European, or Chinese samples. Agarase activity was also identified in the non-marine bacterium Cellvibrio sp. |
| cd06542 | GH18_EndoS-like | 1.34e-04 | 79 | 296 | 19 | 213 | Endo-beta-N-acetylglucosaminidases are bacterial chitinases that hydrolyze the chitin core of various asparagine (N)-linked glycans and glycoproteins. The endo-beta-N-acetylglucosaminidases have a glycosyl hydrolase family 18 (GH18) catalytic domain. Some members also have an additional C-terminal glycosyl hydrolase family 20 (GH20) domain while others have an N-terminal domain of unknown function (pfam08522). Members of this family include endo-beta-N-acetylglucosaminidase S (EndoS) from Streptococcus pyogenes, EndoF1, EndoF2, EndoF3, and EndoH from Flavobacterium meningosepticum, and EndoE from Enterococcus faecalis. EndoS is a secreted endoglycosidase from Streptococcus pyogenes that specifically hydrolyzes the glycan on human IgG between two core N-acetylglucosamine residues. EndoE is a secreted endoglycosidase, encoded by the ndoE gene in Enterococcus faecalis, that hydrolyzes the glycan on human RNase B. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA22857.1 | 4.17e-256 | 4 | 469 | 89 | 552 |
| AVM46100.1 | 8.83e-118 | 34 | 441 | 2 | 410 |
| AVM46102.1 | 5.32e-91 | 42 | 441 | 15 | 409 |
| QEL16017.1 | 7.77e-76 | 31 | 466 | 24 | 465 |
| QJD85864.1 | 4.05e-72 | 32 | 463 | 2 | 445 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5Z3K_A | 1.18e-35 | 32 | 399 | 7 | 368 | Crystalstructure of glucosidase from Croceicoccus marinus at 1.8 Angstrom resolution [Croceicoccus marinus],5Z3K_B Crystal structure of glucosidase from Croceicoccus marinus at 1.8 Angstrom resolution [Croceicoccus marinus] |
| 6UQJ_A | 7.70e-11 | 154 | 292 | 134 | 286 | Crystalstructure of the GH39 enzyme from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
| 4EKJ_A | 9.56e-10 | 156 | 292 | 131 | 281 | ChainA, Beta-xylosidase [Caulobacter vibrioides] |
| 4M29_A | 9.56e-10 | 156 | 292 | 131 | 281 | Structureof a GH39 Beta-xylosidase from Caulobacter crescentus [Caulobacter vibrioides CB15] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23552 | 3.31e-12 | 121 | 346 | 105 | 317 | Beta-xylosidase OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=xynB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
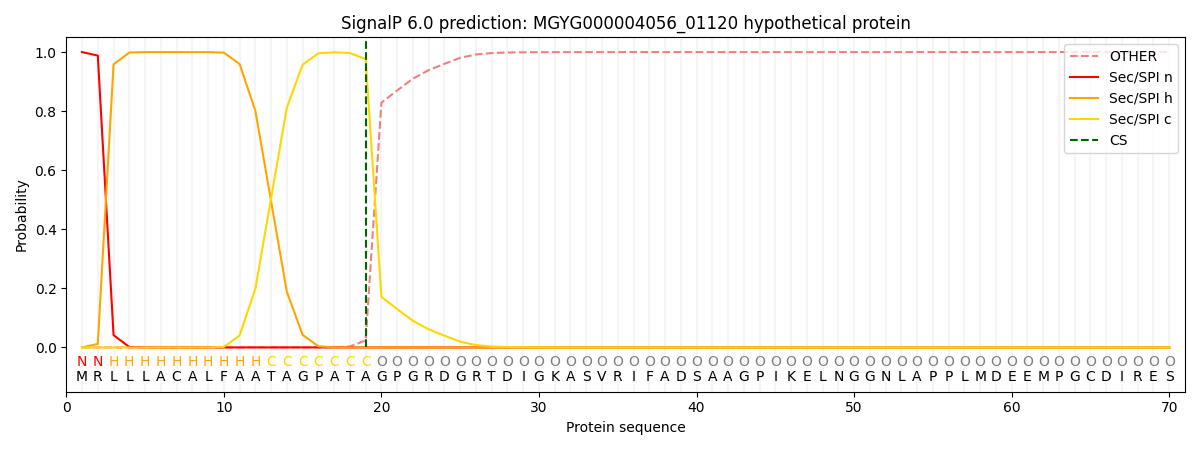
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000258 | 0.999131 | 0.000160 | 0.000171 | 0.000151 | 0.000139 |
