You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004082_00906
You are here: Home > Sequence: MGYG000004082_00906
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | AM51-8 sp900761925 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; AM51-8; AM51-8 sp900761925 | |||||||||||
| CAZyme ID | MGYG000004082_00906 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30423; End: 32534 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 568 | 694 | 2.3e-17 | 0.9296875 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08481 | GBS_Bsp-like | 3.55e-23 | 138 | 217 | 2 | 82 | GBS Bsp-like repeat. This domain is found as a repeat in a number of Streptococcus proteins including some hypothetical proteins and Bsp. Bsp is a protein of group B Streptococcus (GBS) which might control cell morphology. |
| pfam08481 | GBS_Bsp-like | 7.26e-15 | 336 | 411 | 3 | 79 | GBS Bsp-like repeat. This domain is found as a repeat in a number of Streptococcus proteins including some hypothetical proteins and Bsp. Bsp is a protein of group B Streptococcus (GBS) which might control cell morphology. |
| pfam01832 | Glucosaminidase | 1.11e-10 | 568 | 635 | 1 | 77 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| pfam08481 | GBS_Bsp-like | 4.38e-09 | 258 | 319 | 24 | 88 | GBS Bsp-like repeat. This domain is found as a repeat in a number of Streptococcus proteins including some hypothetical proteins and Bsp. Bsp is a protein of group B Streptococcus (GBS) which might control cell morphology. |
| pfam08481 | GBS_Bsp-like | 8.98e-06 | 438 | 518 | 6 | 89 | GBS Bsp-like repeat. This domain is found as a repeat in a number of Streptococcus proteins including some hypothetical proteins and Bsp. Bsp is a protein of group B Streptococcus (GBS) which might control cell morphology. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEN97541.1 | 5.24e-116 | 39 | 694 | 1015 | 1778 |
| QRO35993.1 | 1.88e-99 | 60 | 699 | 1010 | 1762 |
| QBF76227.1 | 1.88e-99 | 60 | 699 | 1010 | 1762 |
| QEI32622.1 | 3.31e-94 | 28 | 692 | 516 | 1302 |
| QRT31212.1 | 3.31e-94 | 28 | 692 | 516 | 1302 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O51481 | 6.94e-19 | 537 | 700 | 47 | 196 | Uncharacterized protein BB_0531 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0531 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
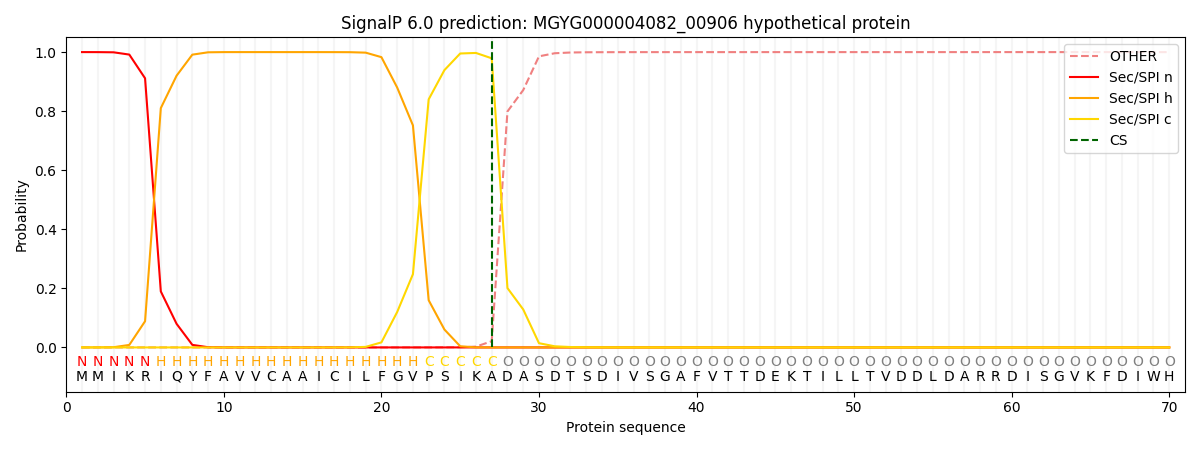
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000319 | 0.998942 | 0.000228 | 0.000183 | 0.000167 | 0.000146 |
