You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004085_00066
You are here: Home > Sequence: MGYG000004085_00066
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UMGS1253; UMGS1253; | |||||||||||
| CAZyme ID | MGYG000004085_00066 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 73352; End: 76516 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT39 | 627 | 854 | 1.9e-56 | 0.9282511210762332 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16192 | PMT_4TMC | 4.14e-29 | 870 | 1049 | 7 | 197 | C-terminal four TMM region of protein-O-mannosyltransferase. PMT_4TMC is the C-terminal four membrane-pass region of protein-O-mannosyltransferases and similar enzymes. |
| COG5542 | COG5542 | 3.97e-20 | 68 | 378 | 71 | 369 | Mannosyltransferase related to Gpi18 [Carbohydrate transport and metabolism]. |
| COG1928 | PMT1 | 5.21e-20 | 858 | 1054 | 493 | 695 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| COG1928 | PMT1 | 4.81e-18 | 630 | 915 | 46 | 323 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| pfam13231 | PMT_2 | 1.34e-13 | 657 | 801 | 2 | 137 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCN30459.1 | 6.48e-265 | 33 | 1053 | 35 | 1057 |
| ASA25569.1 | 6.10e-237 | 9 | 1054 | 221 | 1268 |
| QNM01664.1 | 3.88e-234 | 35 | 1053 | 30 | 1084 |
| AIQ15817.1 | 8.36e-230 | 37 | 1054 | 8 | 1031 |
| AIQ66842.1 | 2.61e-228 | 37 | 1054 | 8 | 1031 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P9WN04 | 2.98e-31 | 634 | 1052 | 73 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=pmt PE=3 SV=2 |
| P9WN05 | 2.98e-31 | 634 | 1052 | 73 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=pmt PE=1 SV=2 |
| Q8NRZ6 | 1.25e-30 | 657 | 1052 | 99 | 519 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=pmt PE=3 SV=1 |
| L8F4Z2 | 2.32e-27 | 634 | 1052 | 67 | 515 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycolicibacterium smegmatis (strain MKD8) OX=1214915 GN=pmt PE=3 SV=1 |
| O67270 | 7.69e-06 | 650 | 773 | 47 | 174 | Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000018 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
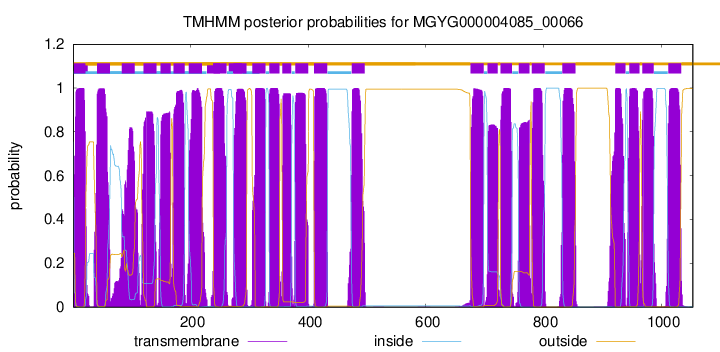
| start | end |
|---|---|
| 2 | 21 |
| 41 | 63 |
| 83 | 105 |
| 120 | 142 |
| 149 | 166 |
| 171 | 190 |
| 197 | 219 |
| 239 | 261 |
| 273 | 295 |
| 305 | 327 |
| 334 | 351 |
| 356 | 371 |
| 378 | 400 |
| 410 | 432 |
| 474 | 496 |
| 676 | 698 |
| 705 | 722 |
| 727 | 746 |
| 758 | 776 |
| 780 | 802 |
| 832 | 854 |
| 922 | 939 |
| 946 | 963 |
| 968 | 987 |
| 1012 | 1034 |
