You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004087_01137
You are here: Home > Sequence: MGYG000004087_01137
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Enterocloster sp900540675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Enterocloster; Enterocloster sp900540675 | |||||||||||
| CAZyme ID | MGYG000004087_01137 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 81884; End: 84694 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 271 | 703 | 1.8e-65 | 0.956140350877193 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 1.19e-57 | 268 | 683 | 3 | 323 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| COG4409 | NanH | 1.63e-21 | 272 | 683 | 268 | 700 | Neuraminidase (sialidase) [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| NF033838 | PspC_subgroup_1 | 5.89e-15 | 730 | 918 | 368 | 572 | pneumococcal surface protein PspC, choline-binding form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. The other form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. |
| COG5263 | COG5263 | 6.44e-15 | 816 | 934 | 209 | 313 | Glucan-binding domain (YG repeat) [Carbohydrate transport and metabolism]. |
| NF033930 | pneumo_PspA | 9.67e-13 | 809 | 918 | 430 | 529 | pneumococcal surface protein A. The pneumococcal surface protein proteins, found in Streptococcus pneumoniae, are repetitive, with patterns of localized high sequence identity across pairs of proteins given different specific names that recombination may be presumed. This protein, PspA, has an N-terminal region that lacks a cross-wall-targeting YSIRK type extended signal peptide, in contrast to the closely related choline-binding protein CbpA which has a similar C-terminus but a YSIRK-containing region at the N-terminus. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARE59829.1 | 3.45e-132 | 243 | 703 | 16 | 510 |
| QQR06511.1 | 3.45e-132 | 243 | 703 | 16 | 510 |
| ANU47084.1 | 2.64e-67 | 259 | 683 | 1202 | 1659 |
| QQQ98205.1 | 2.64e-67 | 259 | 683 | 1202 | 1659 |
| QGN31358.1 | 5.41e-47 | 217 | 687 | 191 | 668 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4X6K_A | 1.02e-44 | 276 | 687 | 16 | 460 | Crystalstructure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with Siastatin B [[Ruminococcus] gnavus CC55_001C] |
| 4X47_A | 1.08e-44 | 276 | 687 | 19 | 463 | Crystalstructure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with Neu5Ac2en [[Ruminococcus] gnavus ATCC 29149],4X49_A Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with oseltamivir carboxylate [[Ruminococcus] gnavus ATCC 29149],4X4A_A Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with 2,7-Anhydro-Neu5Ac [[Ruminococcus] gnavus ATCC 29149] |
| 4YW1_A | 1.34e-43 | 275 | 687 | 203 | 633 | ChainA, Neuraminidase C [Streptococcus pneumoniae TIGR4],4YW1_B Chain B, Neuraminidase C [Streptococcus pneumoniae TIGR4],4YW2_A Chain A, Neuraminidase C [Streptococcus pneumoniae TIGR4],4YW2_B Chain B, Neuraminidase C [Streptococcus pneumoniae TIGR4],4YW3_A Chain A, Neuraminidase C [Streptococcus pneumoniae TIGR4],4YW3_B Chain B, Neuraminidase C [Streptococcus pneumoniae TIGR4],4YW4_A Streptococcus pneumoniae sialidase NanC [Streptococcus pneumoniae],4YW4_B Streptococcus pneumoniae sialidase NanC [Streptococcus pneumoniae],4YW5_A Chain A, Neuraminidase C [Streptococcus pneumoniae TIGR4],4YW5_B Chain B, Neuraminidase C [Streptococcus pneumoniae TIGR4],5F9T_A Chain A, Neuraminidase C [Streptococcus pneumoniae TIGR4],5F9T_B Chain B, Neuraminidase C [Streptococcus pneumoniae TIGR4] |
| 4YZ1_A | 1.63e-43 | 275 | 687 | 222 | 652 | CrystalStructure of Streptococcus pneumoniae NanC, apo structure. [Streptococcus pneumoniae TIGR4],4YZ1_B Crystal Structure of Streptococcus pneumoniae NanC, apo structure. [Streptococcus pneumoniae TIGR4],4YZ2_A Crystal Structure of Streptococcus pneumoniae NanC, in complex with 2-deoxy-2,3-didehydro-N-acetylneuraminic acid. [Streptococcus pneumoniae],4YZ2_B Crystal Structure of Streptococcus pneumoniae NanC, in complex with 2-deoxy-2,3-didehydro-N-acetylneuraminic acid. [Streptococcus pneumoniae],4YZ3_A Crystal Structure of Streptococcus pneumoniae NanC, in complex with Oseltamivir. [Streptococcus pneumoniae TIGR4],4YZ3_B Crystal Structure of Streptococcus pneumoniae NanC, in complex with Oseltamivir. [Streptococcus pneumoniae TIGR4],4YZ4_A Crystal Structure of Streptococcus pneumoniae NanC, in complex with N-Acetylneuraminic acid. [Streptococcus pneumoniae],4YZ4_B Crystal Structure of Streptococcus pneumoniae NanC, in complex with N-Acetylneuraminic acid. [Streptococcus pneumoniae],4YZ5_A Crystal Structure of Streptococcus pneumoniae NanC, in complex with 3-Sialyllactose [Streptococcus pneumoniae],4YZ5_B Crystal Structure of Streptococcus pneumoniae NanC, in complex with 3-Sialyllactose [Streptococcus pneumoniae] |
| 1SLI_A | 1.85e-37 | 276 | 687 | 209 | 653 | LeechIntramolecular Trans-Sialidase Complexed With Dana [Macrobdella decora],1SLL_A Sialidase L From Leech Macrobdella Decora [Macrobdella decora],2SLI_A Leech Intramolecular Trans-Sialidase Complexed With 2,7- Anhydro-Neu5ac, The Reaction Product [Macrobdella decora],3SLI_A Leech Intramolecular Trans-Sialidase Complexed With 2,7- Anhydro-Neu5ac Prepared By Soaking With 3'-Sialyllactose [Macrobdella decora],4SLI_A Leech Intramolecular Trans-Sialidase Complexed With 2- Propenyl-Neu5ac, An Inactive Substrate Analogue [Macrobdella decora] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q27701 | 1.75e-36 | 276 | 687 | 289 | 733 | Anhydrosialidase OS=Macrobdella decora OX=6405 PE=1 SV=1 |
| Q54727 | 1.00e-30 | 279 | 687 | 244 | 673 | Sialidase B OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=nanB PE=1 SV=2 |
| P29767 | 1.31e-30 | 269 | 687 | 385 | 813 | Sialidase OS=Clostridium septicum OX=1504 PE=3 SV=1 |
| P62575 | 5.32e-30 | 275 | 711 | 342 | 791 | Sialidase A OS=Streptococcus pneumoniae OX=1313 GN=nanA PE=1 SV=1 |
| P62576 | 5.32e-30 | 275 | 711 | 342 | 791 | Sialidase A OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=nanA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
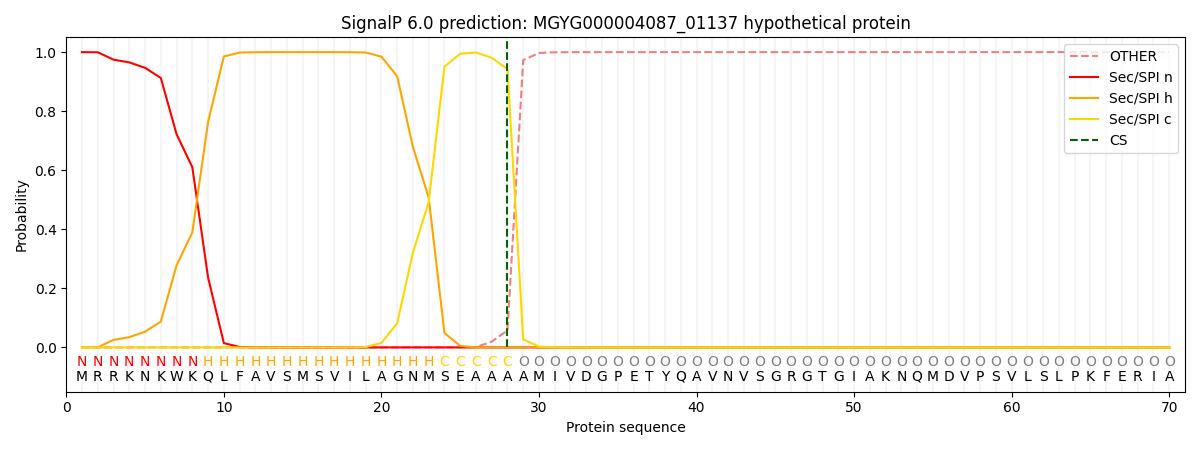
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000386 | 0.998648 | 0.000381 | 0.000199 | 0.000192 | 0.000158 |
