You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004094_00748
You are here: Home > Sequence: MGYG000004094_00748
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; ; | |||||||||||
| CAZyme ID | MGYG000004094_00748 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 29293; End: 30792 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 67 | 499 | 2.1e-164 | 0.9832134292565947 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5520 | XynC | 3.71e-63 | 67 | 499 | 36 | 428 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam02055 | Glyco_hydro_30 | 9.15e-39 | 80 | 409 | 3 | 347 | Glycosyl hydrolase family 30 TIM-barrel domain. |
| pfam02057 | Glyco_hydro_59 | 3.07e-11 | 79 | 367 | 2 | 255 | Glycosyl hydrolase family 59. |
| pfam17189 | Glyco_hydro_30C | 4.54e-09 | 413 | 497 | 1 | 63 | Glycosyl hydrolase family 30 beta sandwich domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOY86583.1 | 1.92e-221 | 10 | 499 | 5 | 497 |
| AHW60617.1 | 1.02e-216 | 6 | 499 | 4 | 501 |
| BBL01056.1 | 4.40e-216 | 17 | 499 | 8 | 491 |
| ATL48908.1 | 6.56e-213 | 10 | 498 | 3 | 488 |
| ADF53788.1 | 1.39e-207 | 28 | 499 | 10 | 481 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5NGK_A | 2.52e-82 | 52 | 499 | 53 | 490 | Theendo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGK_B The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGK_C The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_A The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_B The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_C The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron] |
| 2WNW_A | 7.77e-44 | 39 | 499 | 8 | 442 | Thecrystal structure of SrfJ from salmonella typhimurium [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],2WNW_B The crystal structure of SrfJ from salmonella typhimurium [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 6M5Z_A | 3.63e-16 | 64 | 478 | 22 | 407 | ChainA, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612],6M5Z_B Chain B, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612] |
| 1NOF_A | 3.32e-14 | 170 | 487 | 71 | 340 | ChainA, xylanase [Dickeya chrysanthemi],2Y24_A Chain A, XYLANASE [Dickeya chrysanthemi] |
| 5CXP_A | 1.90e-10 | 177 | 498 | 82 | 386 | X-raycrystallographic protein structure of the glycoside hydrolase family 30 subfamily 8 xylanase, Xyn30A, from Clostridium acetobutylicum [Clostridium acetobutylicum ATCC 824] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4WBR2 | 3.72e-54 | 67 | 423 | 60 | 427 | Endo-1,6-beta-D-glucanase neg1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=neg1 PE=1 SV=1 |
| Q7M4T0 | 6.26e-52 | 55 | 423 | 36 | 416 | Endo-1,6-beta-D-glucanase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=neg-1 PE=1 SV=2 |
| Q8J0I9 | 3.99e-50 | 61 | 423 | 53 | 429 | Endo-1,6-beta-D-glucanase BGN16.3 OS=Trichoderma harzianum OX=5544 PE=1 SV=1 |
| Q9UB00 | 2.16e-35 | 67 | 429 | 88 | 465 | Putative glucosylceramidase 4 OS=Caenorhabditis elegans OX=6239 GN=gba-4 PE=3 SV=2 |
| O16581 | 5.72e-33 | 67 | 468 | 87 | 479 | Putative glucosylceramidase 2 OS=Caenorhabditis elegans OX=6239 GN=gba-2 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
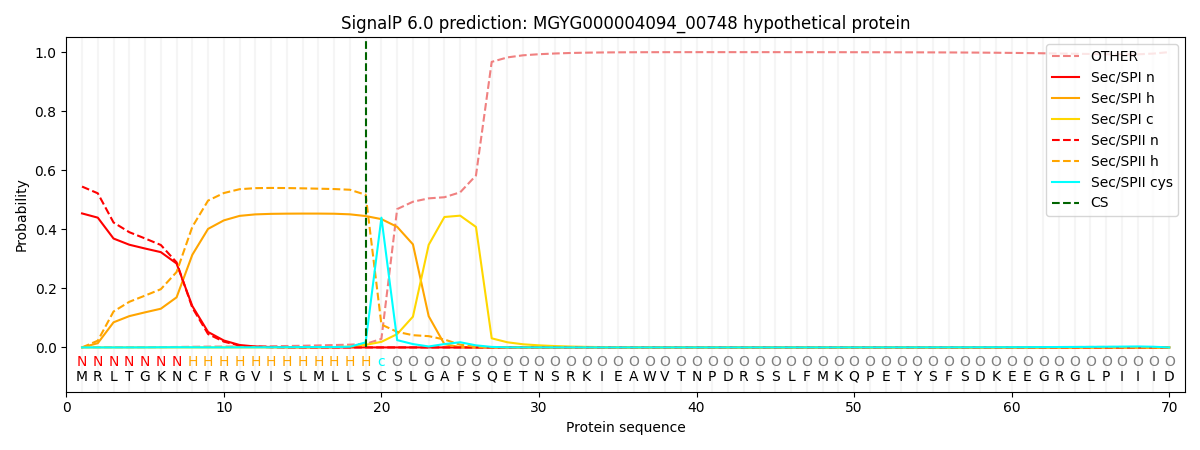
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002789 | 0.436385 | 0.557742 | 0.001760 | 0.000714 | 0.000578 |
