You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004094_00971
You are here: Home > Sequence: MGYG000004094_00971
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; ; | |||||||||||
| CAZyme ID | MGYG000004094_00971 | |||||||||||
| CAZy Family | PL37 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 115043; End: 116914 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL37 | 17 | 614 | 4.7e-193 | 0.9511811023622048 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd02896 | complement_C3_C4_C5 | 0.002 | 99 | 161 | 110 | 181 | Proteins similar to C3, C4 and C5 of vertebrate complement. The vertebrate complement system, comprised of a large number of distinct plasma proteins, is an effector of both the acquired and innate immune systems. The point of convergence of the classical, alternative and lectin pathways of the complement system is the proteolytic activation of C3. C4 plays a key role in propagating the classical and lectin pathways. C5 participates in the classical and alternative pathways. The thioester bond located within the structure of C3 and C4 is central to the function of complement. C5 does not contain an active thioester bond. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANE51487.1 | 5.14e-208 | 2 | 623 | 5 | 625 |
| ACU05303.1 | 1.28e-201 | 5 | 623 | 3 | 628 |
| AXY74233.1 | 8.31e-199 | 1 | 621 | 1 | 620 |
| ACU05681.1 | 2.71e-102 | 3 | 614 | 10 | 627 |
| SDR67806.1 | 2.27e-89 | 34 | 614 | 36 | 619 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KMH5 | 7.25e-63 | 55 | 602 | 66 | 622 | Broad-specificity ulvan lyase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22180 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
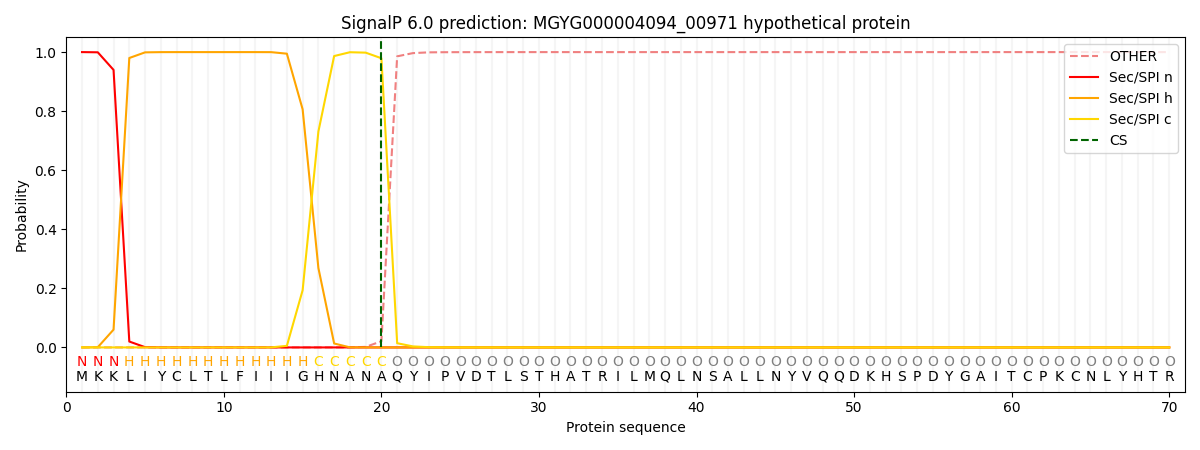
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000244 | 0.999168 | 0.000163 | 0.000152 | 0.000147 | 0.000143 |
