You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004094_01570
You are here: Home > Sequence: MGYG000004094_01570
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; ; | |||||||||||
| CAZyme ID | MGYG000004094_01570 | |||||||||||
| CAZy Family | GH50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 49009; End: 50955 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH50 | 139 | 620 | 1.3e-90 | 0.8315467075038285 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AXP07832.1 | 0.0 | 15 | 648 | 16 | 645 |
| BBE17136.1 | 3.40e-309 | 12 | 648 | 40 | 667 |
| SCM56829.1 | 2.74e-305 | 11 | 646 | 10 | 644 |
| AHF13634.1 | 3.57e-277 | 29 | 647 | 30 | 646 |
| ASB49335.1 | 1.85e-267 | 5 | 648 | 3 | 646 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4BQ2_A | 5.65e-56 | 45 | 626 | 56 | 750 | Structuralanalysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_C Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_D Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_A Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_C Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_D Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40] |
| 4BQ4_A | 1.45e-55 | 45 | 626 | 56 | 750 | Structuralanalysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ4_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ5_A Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ5_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40] |
| 6XJ9_A | 5.40e-54 | 16 | 619 | 50 | 759 | Structureof PfGH50B [Pseudoalteromonas fuliginea],6XJ9_B Structure of PfGH50B [Pseudoalteromonas fuliginea] |
| 5Z6P_A | 2.37e-50 | 178 | 619 | 223 | 759 | Thecrystal structure of an agarase, AgWH50C [Agarivorans gilvus],5Z6P_B The crystal structure of an agarase, AgWH50C [Agarivorans gilvus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P48840 | 9.50e-46 | 125 | 619 | 383 | 949 | Beta-agarase B OS=Vibrio sp. (strain JT0107) OX=47913 GN=agaB PE=3 SV=1 |
| P48839 | 5.67e-32 | 179 | 635 | 400 | 956 | Beta-agarase A OS=Vibrio sp. (strain JT0107) OX=47913 GN=agaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
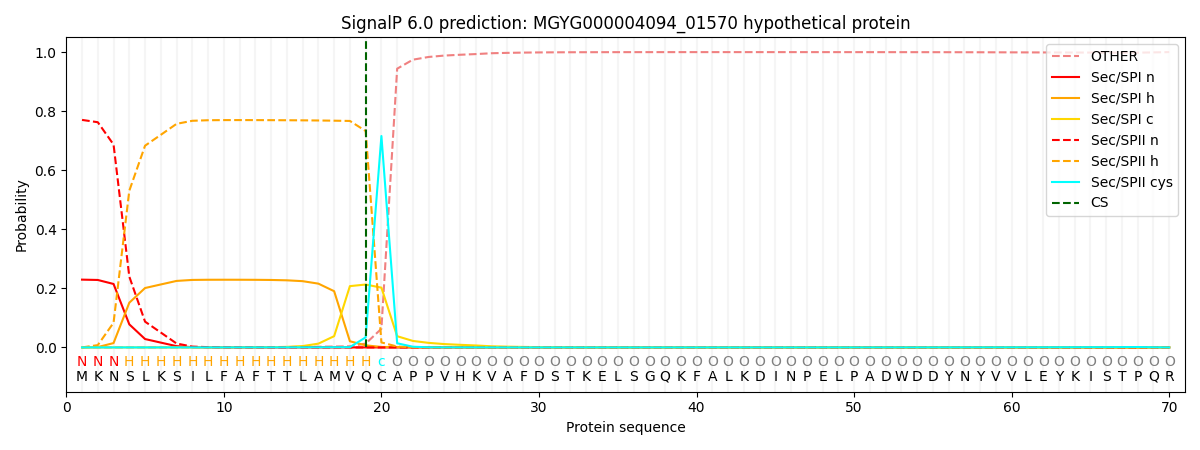
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000714 | 0.223195 | 0.775659 | 0.000134 | 0.000152 | 0.000130 |
