You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004105_01323
You are here: Home > Sequence: MGYG000004105_01323
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes_A sp900546005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes_A; Alistipes_A sp900546005 | |||||||||||
| CAZyme ID | MGYG000004105_01323 | |||||||||||
| CAZy Family | GT22 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 159612; End: 161156 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 18 | 387 | 4.9e-52 | 0.9177377892030848 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03901 | Glyco_transf_22 | 1.06e-20 | 32 | 368 | 20 | 356 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
| PLN02816 | PLN02816 | 3.29e-17 | 6 | 328 | 34 | 350 | mannosyltransferase |
| COG1807 | ArnT | 9.58e-08 | 158 | 359 | 146 | 365 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL06999.1 | 1.35e-144 | 5 | 503 | 9 | 508 |
| AOW16884.1 | 3.24e-67 | 13 | 335 | 11 | 328 |
| AUR51123.1 | 3.78e-65 | 14 | 379 | 12 | 380 |
| QVY67119.1 | 2.32e-62 | 11 | 376 | 9 | 368 |
| QHS61266.1 | 1.23e-59 | 19 | 342 | 8 | 329 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q94A15 | 7.02e-16 | 5 | 328 | 33 | 350 | Mannosyltransferase APTG1 OS=Arabidopsis thaliana OX=3702 GN=APTG1 PE=2 SV=1 |
| Q9VZM5 | 5.26e-15 | 25 | 338 | 18 | 321 | GPI mannosyltransferase 3 OS=Drosophila melanogaster OX=7227 GN=PIG-B PE=2 SV=2 |
| Q6CAB8 | 2.24e-11 | 15 | 363 | 11 | 354 | GPI mannosyltransferase 3 OS=Yarrowia lipolytica (strain CLIB 122 / E 150) OX=284591 GN=GPI10 PE=3 SV=1 |
| Q9JJQ0 | 9.13e-10 | 34 | 392 | 73 | 418 | GPI mannosyltransferase 3 OS=Mus musculus OX=10090 GN=Pigb PE=2 SV=2 |
| Q6BH65 | 1.14e-08 | 31 | 324 | 63 | 355 | GPI mannosyltransferase 3 OS=Debaryomyces hansenii (strain ATCC 36239 / CBS 767 / BCRC 21394 / JCM 1990 / NBRC 0083 / IGC 2968) OX=284592 GN=GPI10 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000013 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
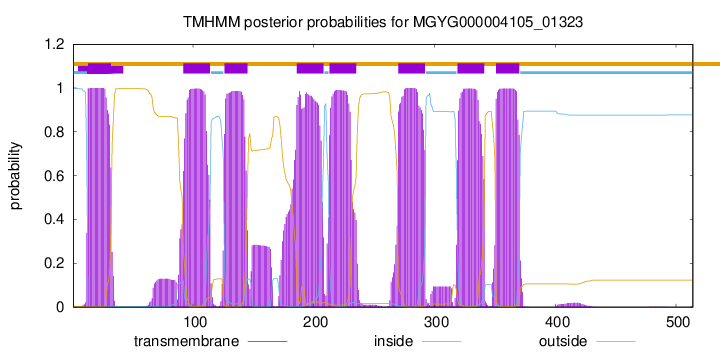
| start | end |
|---|---|
| 13 | 32 |
| 92 | 114 |
| 126 | 145 |
| 186 | 208 |
| 213 | 235 |
| 270 | 292 |
| 319 | 341 |
| 351 | 370 |
