You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004105_01557
You are here: Home > Sequence: MGYG000004105_01557
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes_A sp900546005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes_A; Alistipes_A sp900546005 | |||||||||||
| CAZyme ID | MGYG000004105_01557 | |||||||||||
| CAZy Family | GH78 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30101; End: 32566 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH78 | 98 | 571 | 2.8e-72 | 0.9642857142857143 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17390 | Bac_rhamnosid_C | 2.97e-06 | 523 | 584 | 1 | 64 | Bacterial alpha-L-rhamnosidase C-terminal domain. This family consists of bacterial rhamnosidase A and B enzymes. L-Rhamnose is abundant in biomass as a common constituent of glycolipids and glycosides, such as plant pigments, pectic polysaccharides, gums or biosurfactants. Some rhamnosides are important bioactive compounds. For example, terpenyl glycosides, the glycosidic precursor of aromatic terpenoids, act as important flavouring substances in grapes. Other rhamnosides act as cytotoxic rhamnosylated terpenoids, as signal substances in plants or play a role in the antigenicity of pathogenic bacteria. |
| pfam17389 | Bac_rhamnosid6H | 0.002 | 203 | 517 | 4 | 336 | Bacterial alpha-L-rhamnosidase 6 hairpin glycosidase domain. This family consists of bacterial rhamnosidase A and B enzymes. L-Rhamnose is abundant in biomass as a common constituent of glycolipids and glycosides, such as plant pigments, pectic polysaccharides, gums or biosurfactants. Some rhamnosides are important bioactive compounds. For example, terpenyl glycosides, the glycosidic precursor of aromatic terpenoids, act as important flavouring substances in grapes. Other rhamnosides act as cytotoxic rhamnosylated terpenoids, as signal substances in plants or play a role in the antigenicity of pathogenic bacteria. |
| cd04103 | Centaurin_gamma | 0.006 | 563 | 599 | 8 | 46 | Centaurin gamma (CENTG) GTPase. The centaurins (alpha, beta, gamma, and delta) are large, multi-domain proteins that all contain an ArfGAP domain and ankyrin repeats, and in some cases, numerous additional domains. Centaurin gamma contains an additional GTPase domain near its N-terminus. The specific function of this GTPase domain has not been well characterized, but centaurin gamma 2 (CENTG2) may play a role in the development of autism. Centaurin gamma 1 is also called PIKE (phosphatidyl inositol (PI) 3-kinase enhancer) and centaurin gamma 2 is also known as AGAP (ArfGAP protein with a GTPase-like domain, ankyrin repeats and a Pleckstrin homology domain) or GGAP. Three isoforms of PIKE have been identified. PIKE-S (short) and PIKE-L (long) are brain-specific isoforms, with PIKE-S restricted to the nucleus and PIKE-L found in multiple cellular compartments. A third isoform, PIKE-A was identified in human glioblastoma brain cancers and has been found in various tissues. GGAP has been shown to have high GTPase activity due to a direct intramolecular interaction between the N-terminal GTPase domain and the C-terminal ArfGAP domain. In human tissue, AGAP mRNA was detected in skeletal muscle, kidney, placenta, brain, heart, colon, and lung. Reduced expression levels were also observed in the spleen, liver, and small intestine. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA24541.1 | 0.0 | 11 | 821 | 11 | 820 |
| ATP56756.1 | 1.54e-286 | 36 | 821 | 31 | 818 |
| QMW04815.1 | 1.29e-285 | 31 | 821 | 32 | 816 |
| ACT93515.1 | 1.32e-281 | 33 | 821 | 51 | 831 |
| QEC76313.1 | 2.93e-277 | 36 | 821 | 45 | 844 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CIH_A | 7.73e-36 | 22 | 573 | 100 | 694 | Crystalstructure of a putative alpha-rhamnosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 2OKX_A | 6.45e-17 | 103 | 556 | 439 | 910 | Crystalstructure of GH78 family rhamnosidase of Bacillus SP. GL1 AT 1.9 A [Bacillus sp. GL1],2OKX_B Crystal structure of GH78 family rhamnosidase of Bacillus SP. GL1 AT 1.9 A [Bacillus sp. GL1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P9WF03 | 2.14e-07 | 466 | 628 | 766 | 910 | Alpha-L-rhamnosidase OS=Alteromonas sp. (strain LOR) OX=1537994 GN=LOR_34 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
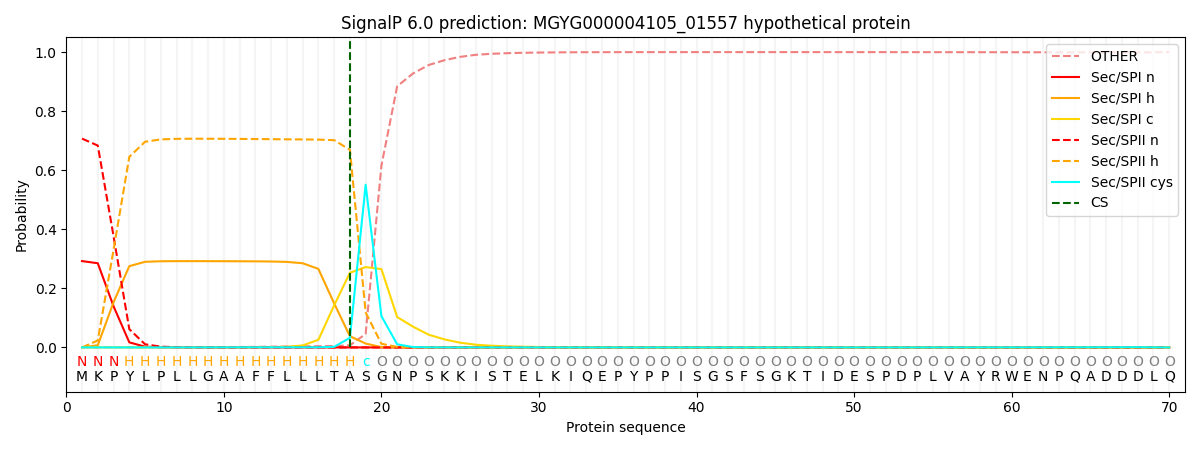
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001675 | 0.286097 | 0.711804 | 0.000155 | 0.000145 | 0.000125 |
