You are browsing environment: HUMAN GUT
MGYG000004116_02930
Basic Information
help
Species
Lineage
Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; UMGS1591;
CAZyme ID
MGYG000004116_02930
CAZy Family
GH110
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000004116
3787047
MAG
United Kingdom
Europe
Gene Location
Start: 23852;
End: 26011
Strand: +
No EC number prediction in MGYG000004116_02930.
Family
Start
End
Evalue
family coverage
GH110
173
654
2.4e-44
0.8284671532846716
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam13229
Beta_helix
4.67e-05
541
715
10
157
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases.
more
pfam05048
NosD
1.32e-04
550
716
9
172
Periplasmic copper-binding protein (NosD). NosD is a periplasmic protein which is thought to insert copper into the exported reductase apoenzyme (NosZ). This region forms a parallel beta helix domain.
more
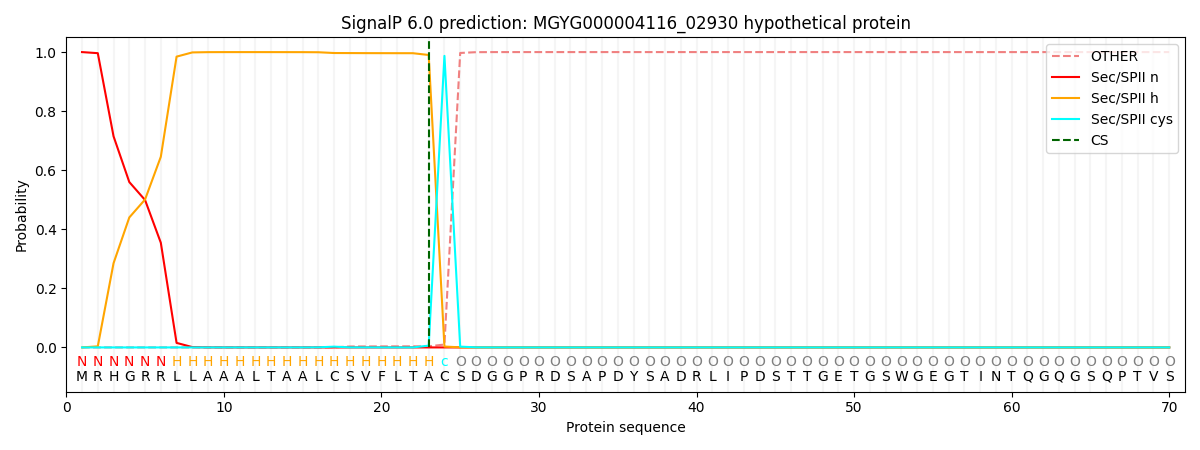
This protein is predicted as LIPO
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000000
0.000001
1.000057
0.000000
0.000000
0.000000
There is no transmembrane helices in MGYG000004116_02930.