You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004128_00209
You are here: Home > Sequence: MGYG000004128_00209
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
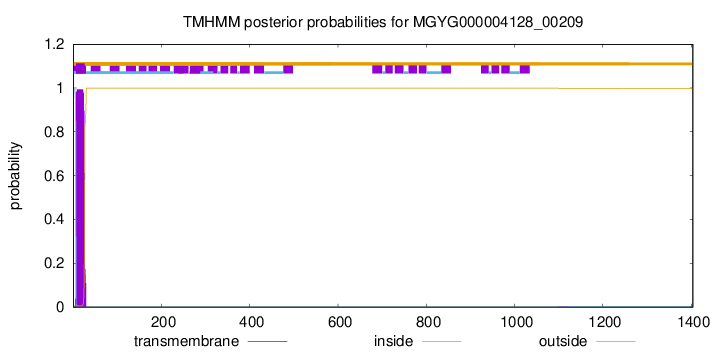
TMHMM annotations
Basic Information help
| Species | UMGS1253 sp900550065 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UMGS1253; UMGS1253; UMGS1253 sp900550065 | |||||||||||
| CAZyme ID | MGYG000004128_00209 | |||||||||||
| CAZy Family | PL21 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 31113; End: 35327 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL21 | 753 | 819 | 9.8e-23 | 0.9722222222222222 |
| CBM32 | 1289 | 1398 | 4e-16 | 0.8467741935483871 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07833 | Cu_amine_oxidN1 | 2.29e-25 | 1158 | 1250 | 1 | 93 | Copper amine oxidase N-terminal domain. Copper amine oxidases catalyze the oxidative deamination of primary amines to the corresponding aldehydes, while reducing molecular oxygen to hydrogen peroxide. These enzymes are dimers of identical subunits, each comprising four domains. The N-terminal domain, which is absent in some amine oxidases, consists of a five-stranded antiparallel beta sheet twisted around an alpha helix. The D1 domains from the two subunits comprise the 'stalk' of the mushroom-shaped dimer, and interact with each other but do not pack tightly against each other. |
| pfam18675 | HepII_C | 7.25e-12 | 1034 | 1119 | 1 | 88 | Heparinase II C-terminal domain. Heparinase II (HepII) is an 85-kDa dimeric enzyme that depolymerizes both heparin and heparan sulfate glycosaminoglycans. The protein is composed of three domains: an N-terminal alpha-helical domain, a central two-layered beta-sheet domain, and a C-terminal domain forming a two-layered beta-sheet. The C-terminal domain contains nine beta-strands packed together in a manner resembling a beta-barrel. |
| pfam00754 | F5_F8_type_C | 1.54e-11 | 1289 | 1398 | 12 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam07833 | Cu_amine_oxidN1 | 3.04e-07 | 1131 | 1183 | 29 | 88 | Copper amine oxidase N-terminal domain. Copper amine oxidases catalyze the oxidative deamination of primary amines to the corresponding aldehydes, while reducing molecular oxygen to hydrogen peroxide. These enzymes are dimers of identical subunits, each comprising four domains. The N-terminal domain, which is absent in some amine oxidases, consists of a five-stranded antiparallel beta sheet twisted around an alpha helix. The D1 domains from the two subunits comprise the 'stalk' of the mushroom-shaped dimer, and interact with each other but do not pack tightly against each other. |
| pfam07833 | Cu_amine_oxidN1 | 6.25e-07 | 1220 | 1251 | 1 | 32 | Copper amine oxidase N-terminal domain. Copper amine oxidases catalyze the oxidative deamination of primary amines to the corresponding aldehydes, while reducing molecular oxygen to hydrogen peroxide. These enzymes are dimers of identical subunits, each comprising four domains. The N-terminal domain, which is absent in some amine oxidases, consists of a five-stranded antiparallel beta sheet twisted around an alpha helix. The D1 domains from the two subunits comprise the 'stalk' of the mushroom-shaped dimer, and interact with each other but do not pack tightly against each other. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI63941.1 | 1.27e-197 | 373 | 1115 | 21 | 787 |
| BCI63940.1 | 7.52e-180 | 389 | 1122 | 41 | 796 |
| BBH20141.1 | 2.42e-179 | 384 | 1389 | 228 | 1210 |
| QOU74174.1 | 1.00e-156 | 217 | 1126 | 31 | 947 |
| AYM77060.1 | 1.43e-155 | 217 | 1126 | 31 | 947 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2FUQ_A | 4.31e-140 | 391 | 1119 | 19 | 747 | ChainA, heparinase II protein [Pedobacter heparinus],2FUQ_B Chain B, heparinase II protein [Pedobacter heparinus] |
| 3E80_A | 4.56e-140 | 391 | 1119 | 21 | 749 | Structureof Heparinase II complexed with heparan sulfate degradation disaccharide product [Pedobacter heparinus],3E80_B Structure of Heparinase II complexed with heparan sulfate degradation disaccharide product [Pedobacter heparinus],3E80_C Structure of Heparinase II complexed with heparan sulfate degradation disaccharide product [Pedobacter heparinus] |
| 3E7J_A | 2.44e-137 | 391 | 1119 | 21 | 749 | ChainA, Heparinase II protein [Pedobacter heparinus],3E7J_B Chain B, Heparinase II protein [Pedobacter heparinus] |
| 2FUT_A | 2.42e-133 | 391 | 1119 | 20 | 748 | ChainA, heparinase II protein [Pedobacter heparinus],2FUT_B Chain B, heparinase II protein [Pedobacter heparinus] |
| 5ZU6_A | 1.47e-18 | 1295 | 1401 | 51 | 157 | ACBM32 derived from alginate lyase B (AlyB-OU02) [Vibrio] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| C6XZB6 | 4.81e-139 | 391 | 1119 | 44 | 772 | Heparin and heparin-sulfate lyase OS=Pedobacter heparinus (strain ATCC 13125 / DSM 2366 / CIP 104194 / JCM 7457 / NBRC 12017 / NCIMB 9290 / NRRL B-14731 / HIM 762-3) OX=485917 GN=hepB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
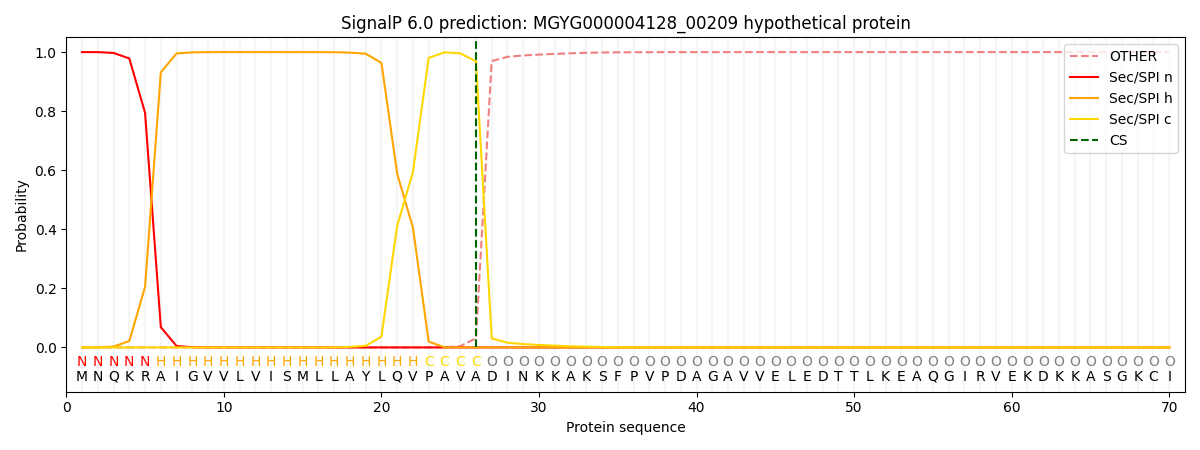
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000342 | 0.998957 | 0.000156 | 0.000202 | 0.000169 | 0.000164 |