You are browsing environment: HUMAN GUT
MGYG000004128_00726
Basic Information
help
Species
UMGS1253 sp900550065
Lineage
Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UMGS1253; UMGS1253; UMGS1253 sp900550065
CAZyme ID
MGYG000004128_00726
CAZy Family
PL35
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
2043
221399.52
4.5063
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000004128
2239270
MAG
United Kingdom
Europe
Gene Location
Start: 7177;
End: 13308
Strand: -
No EC number prediction in MGYG000004128_00726.
Family
Start
End
Evalue
family coverage
PL35
725
903
4e-50
0.9720670391061452
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam16332
DUF4962
3.85e-06
365
617
126
385
Domain of unknown function (DUF4962). This family consists of uncharacterized proteins around 870 residues in length and is mainly found in various Bacteroides species. The function of this protein is unknown.
more
pfam13205
Big_5
4.74e-06
1741
1826
22
106
Bacterial Ig-like domain.
more
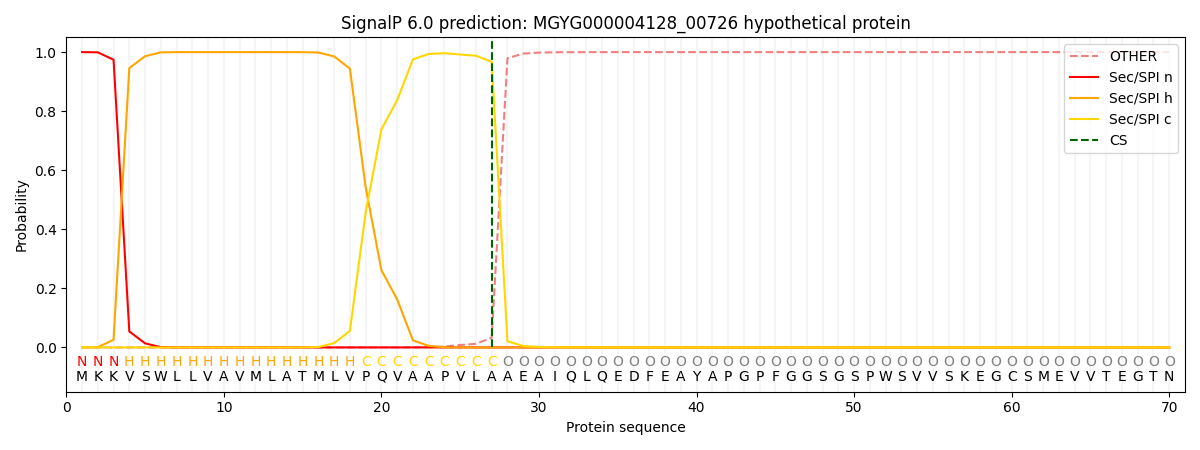
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000226
0.999124
0.000161
0.000164
0.000142
0.000140