You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004128_00767
You are here: Home > Sequence: MGYG000004128_00767
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
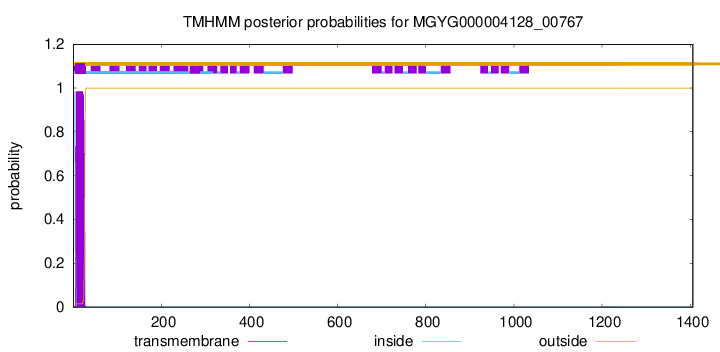
TMHMM annotations
Basic Information help
| Species | UMGS1253 sp900550065 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UMGS1253; UMGS1253; UMGS1253 sp900550065 | |||||||||||
| CAZyme ID | MGYG000004128_00767 | |||||||||||
| CAZy Family | PL35 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16186; End: 20406 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL35 | 926 | 1098 | 1.6e-41 | 0.9608938547486033 |
| CBM32 | 1283 | 1402 | 3.9e-20 | 0.9354838709677419 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07833 | Cu_amine_oxidN1 | 1.94e-26 | 446 | 536 | 1 | 92 | Copper amine oxidase N-terminal domain. Copper amine oxidases catalyze the oxidative deamination of primary amines to the corresponding aldehydes, while reducing molecular oxygen to hydrogen peroxide. These enzymes are dimers of identical subunits, each comprising four domains. The N-terminal domain, which is absent in some amine oxidases, consists of a five-stranded antiparallel beta sheet twisted around an alpha helix. The D1 domains from the two subunits comprise the 'stalk' of the mushroom-shaped dimer, and interact with each other but do not pack tightly against each other. |
| pfam07833 | Cu_amine_oxidN1 | 4.71e-15 | 392 | 476 | 10 | 93 | Copper amine oxidase N-terminal domain. Copper amine oxidases catalyze the oxidative deamination of primary amines to the corresponding aldehydes, while reducing molecular oxygen to hydrogen peroxide. These enzymes are dimers of identical subunits, each comprising four domains. The N-terminal domain, which is absent in some amine oxidases, consists of a five-stranded antiparallel beta sheet twisted around an alpha helix. The D1 domains from the two subunits comprise the 'stalk' of the mushroom-shaped dimer, and interact with each other but do not pack tightly against each other. |
| pfam00754 | F5_F8_type_C | 5.11e-14 | 1282 | 1401 | 3 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam16332 | DUF4962 | 2.42e-07 | 573 | 853 | 133 | 417 | Domain of unknown function (DUF4962). This family consists of uncharacterized proteins around 870 residues in length and is mainly found in various Bacteroides species. The function of this protein is unknown. |
| cd00057 | FA58C | 0.001 | 1283 | 1356 | 16 | 92 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUH30172.1 | 2.37e-150 | 575 | 1399 | 71 | 888 |
| AZS17848.1 | 1.72e-147 | 268 | 1400 | 108 | 1273 |
| ALS27107.1 | 1.65e-127 | 556 | 1402 | 677 | 1509 |
| SDS98549.1 | 2.31e-122 | 577 | 1400 | 52 | 854 |
| QNK58643.1 | 8.74e-120 | 543 | 1400 | 759 | 1605 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5ZU6_A | 3.44e-24 | 1305 | 1404 | 57 | 157 | ACBM32 derived from alginate lyase B (AlyB-OU02) [Vibrio] |
| 5ZU5_A | 1.09e-21 | 1305 | 1404 | 57 | 157 | Crystalstructure of a full length alginate lyase with CBM domain [Vibrio splendidus] |
| 7D29_A | 2.55e-15 | 1300 | 1402 | 29 | 130 | CBM32of AlyQ [Persicobacter sp. CCB-QB2],7D2A_A CBM32 of AlyQ in complex with 4,5-unsaturated mannuronic acid [Persicobacter sp. CCB-QB2] |
| 5XNR_A | 1.28e-13 | 1300 | 1402 | 29 | 130 | TruncatedAlyQ with CBM32 and alginate lyase domains [Persicobacter sp. CCB-QB2] |
| 2JD9_A | 1.32e-11 | 1277 | 1402 | 9 | 132 | Structureof a pectin binding carbohydrate binding module determined in an orthorhombic crystal form. [Yersinia enterocolitica],2JDA_A Structure of a pectin binding carbohydrate binding module determined in an monoclinic crystal form. [Yersinia enterocolitica],2JDA_B Structure of a pectin binding carbohydrate binding module determined in an monoclinic crystal form. [Yersinia enterocolitica] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
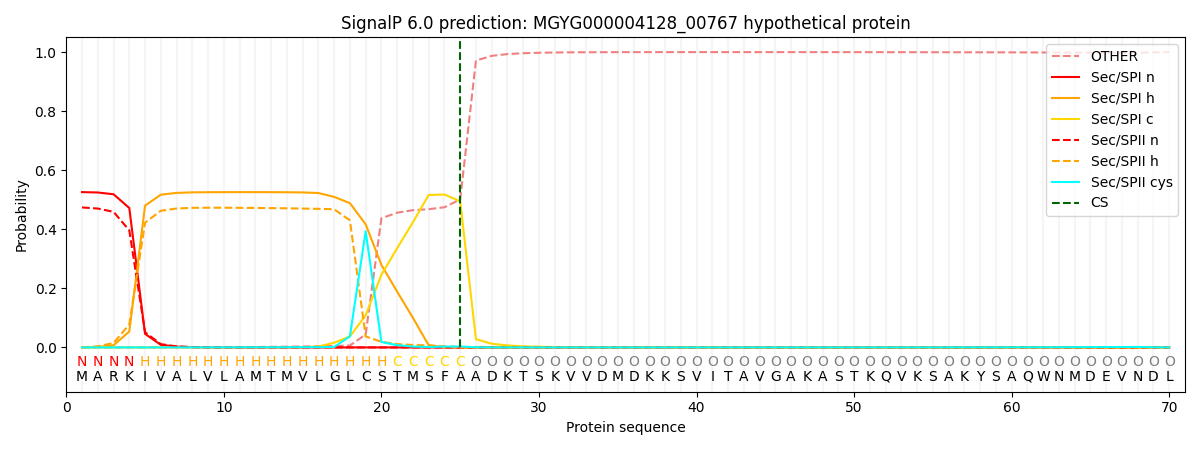
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000721 | 0.516622 | 0.481912 | 0.000314 | 0.000240 | 0.000190 |