You are browsing environment: HUMAN GUT
MGYG000004132_00317
Basic Information
help
Species
V9D3004 sp900760345
Lineage
Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; V9D3004; V9D3004 sp900760345
CAZyme ID
MGYG000004132_00317
CAZy Family
GH95
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000004132
2510246
MAG
United Kingdom
Europe
Gene Location
Start: 94560;
End: 97502
Strand: -
No EC number prediction in MGYG000004132_00317.
Family
Start
End
Evalue
family coverage
GH95
233
717
5.2e-22
0.610803324099723
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam02368
Big_2
6.03e-11
48
113
11
77
Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins.
more
smart00635
BID_2
2.67e-06
36
113
1
81
Bacterial Ig-like domain 2.
more
PRK09537
pylS
0.004
42
159
31
154
pyrrolysine--tRNA(Pyl) ligase.
more
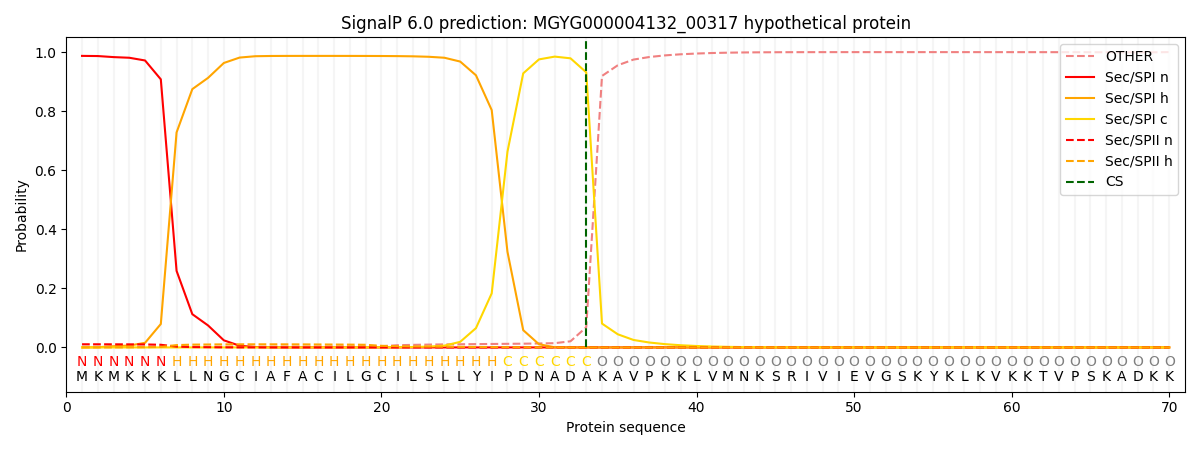
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.004625
0.982350
0.012142
0.000325
0.000263
0.000264