You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004140_00196
You are here: Home > Sequence: MGYG000004140_00196
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
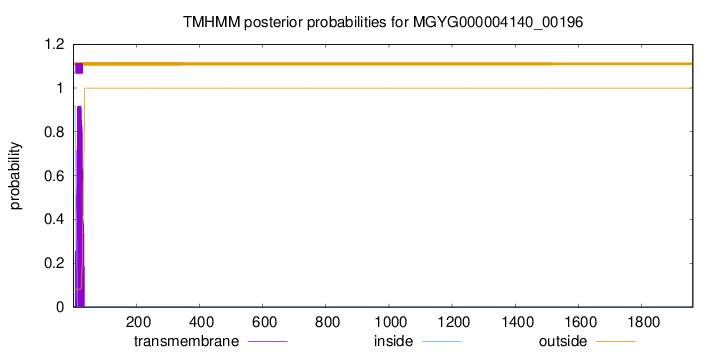
TMHMM annotations
Basic Information help
| Species | UMGS1375 sp900551235 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; UMGS1375; UMGS1375 sp900551235 | |||||||||||
| CAZyme ID | MGYG000004140_00196 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 223001; End: 228892 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 124 | 248 | 4.9e-19 | 0.8629032258064516 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5492 | YjdB | 7.95e-25 | 1466 | 1715 | 99 | 327 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG5492 | YjdB | 2.43e-24 | 1385 | 1624 | 109 | 327 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam00754 | F5_F8_type_C | 2.77e-21 | 110 | 248 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| COG5492 | YjdB | 7.54e-18 | 1599 | 1720 | 140 | 261 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam02368 | Big_2 | 1.29e-15 | 1550 | 1627 | 1 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDO70357.1 | 3.58e-238 | 264 | 1140 | 94 | 917 |
| ALG48827.1 | 5.77e-238 | 4 | 1150 | 8 | 1182 |
| ABG83191.1 | 1.18e-235 | 4 | 1150 | 8 | 1182 |
| AXH52486.1 | 4.46e-235 | 4 | 1150 | 8 | 1182 |
| QUT26415.1 | 7.13e-235 | 264 | 1136 | 94 | 913 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1GOF_A | 1.32e-12 | 104 | 258 | 9 | 153 | NOVELTHIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],1GOG_A NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],1GOH_A NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],2EIE_A Chain A, Galactose oxidase [Fusarium graminearum],2JKX_A Chain A, GALACTOSE OXIDASE [Fusarium graminearum],2VZ1_A Chain A, GALACTOSE OXIDASE [Fusarium graminearum],2VZ3_A Chain A, Galactose Oxidase [Fusarium graminearum] |
| 2EID_A | 1.32e-12 | 104 | 258 | 9 | 153 | ChainA, Galactose oxidase [Fusarium graminearum] |
| 2EIC_A | 1.32e-12 | 104 | 258 | 9 | 153 | ChainA, Galactose oxidase [Fusarium graminearum] |
| 2EIB_A | 1.32e-12 | 104 | 258 | 9 | 153 | ChainA, Galactose oxidase [Fusarium graminearum] |
| 1T2X_A | 1.32e-12 | 104 | 258 | 9 | 153 | Glactoseoxidase C383S mutant identified by directed evolution [Fusarium sp.] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P85991 | 2.18e-18 | 1523 | 1721 | 65 | 258 | Ig-like virion protein OS=Serratia phage KSP90 OX=552528 PE=1 SV=2 |
| P33747 | 1.44e-12 | 1541 | 1721 | 29 | 202 | Uncharacterized protein CA_P0160 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_P0160 PE=3 SV=2 |
| I1S2N3 | 5.83e-12 | 104 | 258 | 50 | 194 | Galactose oxidase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=GAOA PE=3 SV=1 |
| P0CS93 | 7.67e-12 | 104 | 258 | 50 | 194 | Galactose oxidase OS=Gibberella zeae OX=5518 GN=GAOA PE=1 SV=1 |
| Q02834 | 1.18e-08 | 102 | 250 | 502 | 643 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
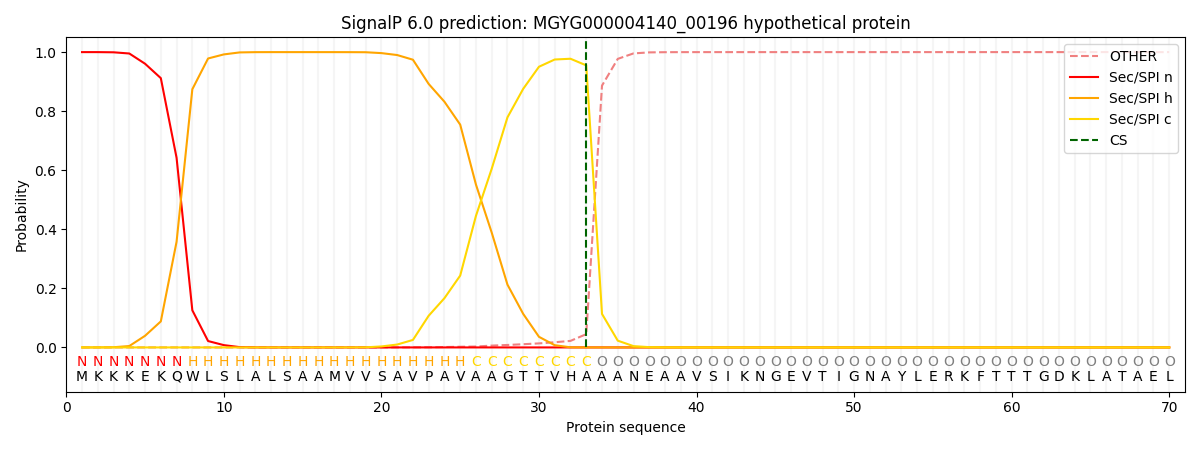
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000405 | 0.998667 | 0.000210 | 0.000267 | 0.000229 | 0.000189 |