You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004168_02347
You are here: Home > Sequence: MGYG000004168_02347
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-74; Firm-11; | |||||||||||
| CAZyme ID | MGYG000004168_02347 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11708; End: 13087 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 178 | 434 | 1.4e-44 | 0.7476923076923077 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 2.48e-13 | 126 | 358 | 141 | 394 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 8.70e-08 | 181 | 406 | 48 | 307 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PLN03003 | PLN03003 | 4.98e-05 | 186 | 291 | 112 | 222 | Probable polygalacturonase At3g15720 |
| PLN02793 | PLN02793 | 0.001 | 160 | 286 | 119 | 256 | Probable polygalacturonase |
| pfam13229 | Beta_helix | 0.002 | 212 | 356 | 3 | 136 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABX43664.1 | 8.13e-120 | 34 | 454 | 81 | 515 |
| BCK00431.1 | 9.90e-113 | 34 | 444 | 83 | 507 |
| ACI19833.1 | 4.98e-111 | 34 | 459 | 49 | 488 |
| QTE67472.1 | 2.34e-108 | 28 | 447 | 43 | 478 |
| ACK41503.1 | 1.31e-107 | 34 | 447 | 49 | 475 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3JUR_A | 1.79e-08 | 212 | 438 | 193 | 417 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
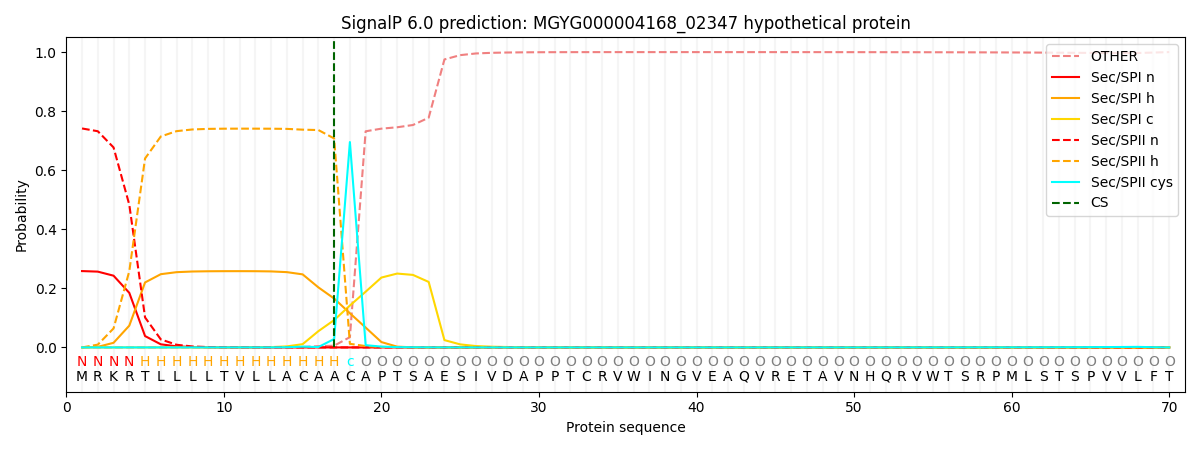
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000466 | 0.251246 | 0.747571 | 0.000336 | 0.000218 | 0.000162 |
