You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004169_01377
You are here: Home > Sequence: MGYG000004169_01377
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
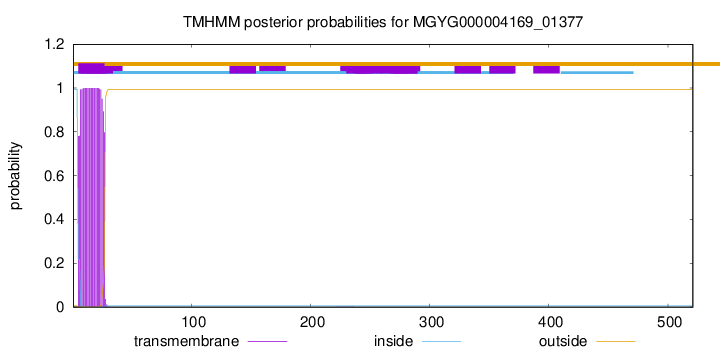
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; UMGS2037; | |||||||||||
| CAZyme ID | MGYG000004169_01377 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1091; End: 2656 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 234 | 507 | 7.5e-39 | 0.7661538461538462 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 1.40e-11 | 132 | 341 | 45 | 315 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 1.30e-05 | 261 | 421 | 81 | 237 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PLN03003 | PLN03003 | 7.89e-05 | 247 | 424 | 119 | 292 | Probable polygalacturonase At3g15720 |
| PLN02793 | PLN02793 | 0.003 | 245 | 345 | 147 | 260 | Probable polygalacturonase |
| smart00557 | IG_FLMN | 0.005 | 124 | 179 | 31 | 80 | Filamin-type immunoglobulin domains. These form a rod-like structure in the actin-binding cytoskeleton protein, filamin. The C-terminal repeats of filamin bind beta1-integrin (CD29). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABX43664.1 | 4.69e-237 | 17 | 521 | 13 | 518 |
| BCK00431.1 | 9.54e-223 | 16 | 521 | 15 | 520 |
| ACI19833.1 | 5.22e-140 | 43 | 510 | 28 | 474 |
| ACK41503.1 | 1.65e-138 | 43 | 510 | 28 | 474 |
| QTE67472.1 | 1.03e-129 | 48 | 512 | 33 | 479 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7PZL3 | 1.07e-08 | 209 | 342 | 136 | 278 | Probable polygalacturonase OS=Vitis vinifera OX=29760 GN=GSVIVT00026920001 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
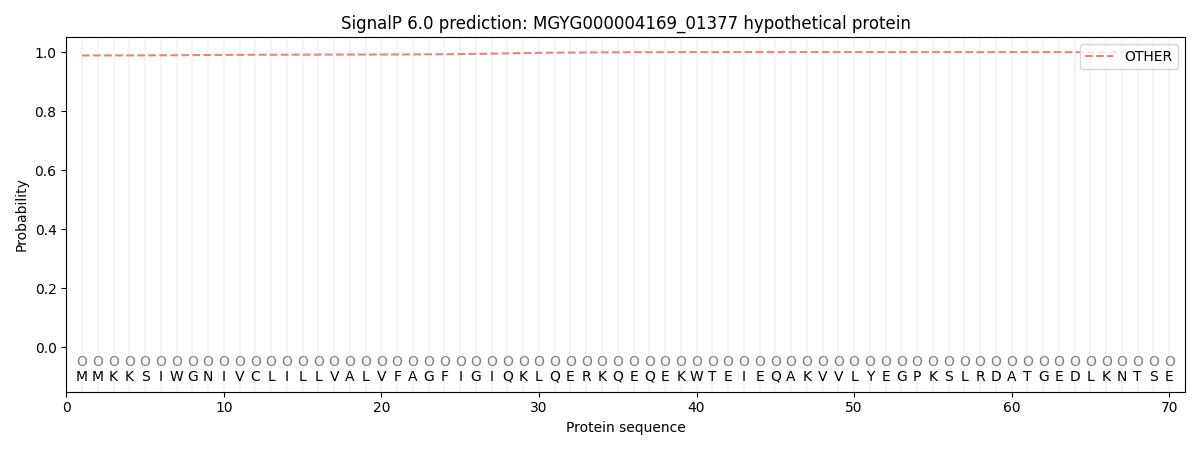
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.989131 | 0.008037 | 0.000838 | 0.000059 | 0.000031 | 0.001942 |