You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004180_01921
You are here: Home > Sequence: MGYG000004180_01921
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
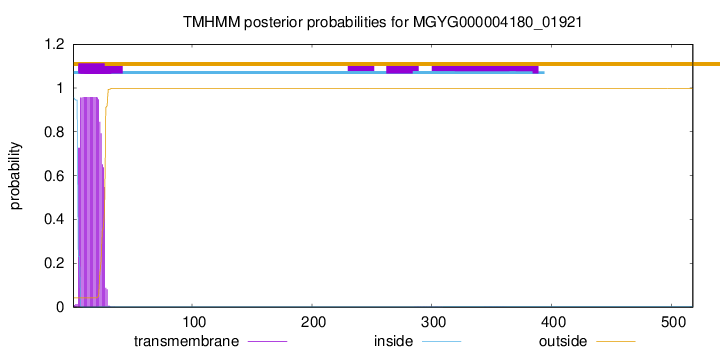
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; | |||||||||||
| CAZyme ID | MGYG000004180_01921 | |||||||||||
| CAZy Family | GH142 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 73711; End: 75267 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH142 | 39 | 512 | 3.4e-167 | 0.9937369519832986 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam06202 | GDE_C | 3.19e-08 | 156 | 348 | 90 | 302 | Amylo-alpha-1,6-glucosidase. This family includes human glycogen branching enzyme AGL. This enzyme contains a number of distinct catalytic activities. It has been shown for the yeast homolog GDB1 that mutations in this region disrupt the enzymes Amylo-alpha-1,6-glucosidase (EC:3.2.1.33). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT49320.1 | 0.0 | 1 | 518 | 1 | 518 |
| QRO14849.1 | 6.24e-262 | 9 | 518 | 8 | 515 |
| AST55953.1 | 6.24e-262 | 9 | 518 | 8 | 515 |
| QUT20767.1 | 6.24e-262 | 9 | 518 | 8 | 515 |
| QUT97167.1 | 6.24e-262 | 9 | 518 | 8 | 515 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQS_A | 8.39e-108 | 35 | 517 | 629 | 1104 | SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 5MQR_A | 9.04e-101 | 35 | 517 | 629 | 1104 | SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
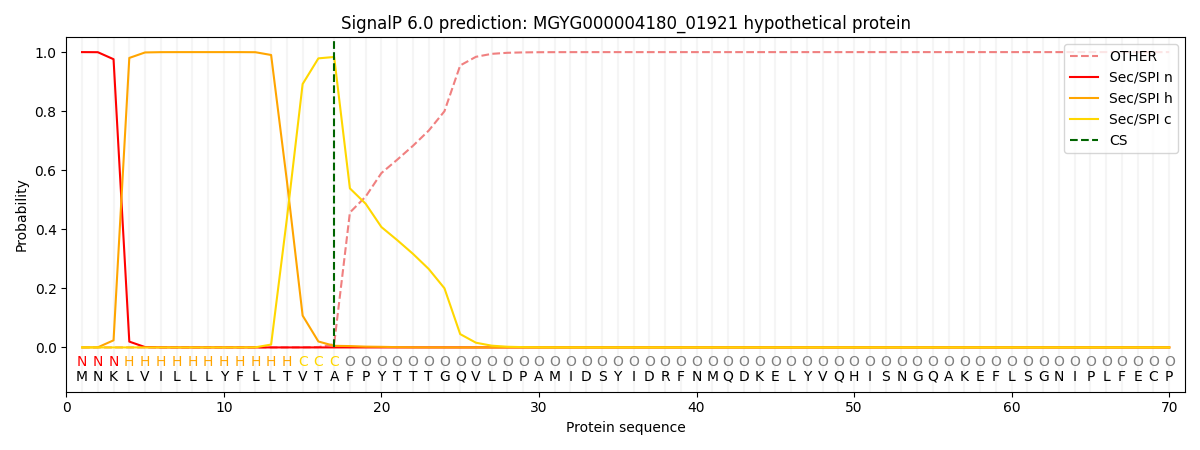
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000220 | 0.999153 | 0.000143 | 0.000166 | 0.000145 | 0.000140 |