You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004182_00507
You are here: Home > Sequence: MGYG000004182_00507
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp900551445 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900551445 | |||||||||||
| CAZyme ID | MGYG000004182_00507 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 61721; End: 63289 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 176 | 472 | 5e-92 | 0.9891304347826086 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 4.89e-50 | 173 | 472 | 13 | 269 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.29e-19 | 146 | 518 | 45 | 407 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam13205 | Big_5 | 1.33e-06 | 31 | 127 | 5 | 106 | Bacterial Ig-like domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIK55671.1 | 1.49e-103 | 121 | 502 | 113 | 478 |
| QIK61079.1 | 8.33e-103 | 121 | 472 | 113 | 459 |
| ADQ18612.1 | 1.09e-100 | 120 | 516 | 176 | 551 |
| BAF57333.1 | 2.27e-95 | 141 | 512 | 20 | 366 |
| ASB50110.1 | 3.81e-89 | 136 | 516 | 220 | 573 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OYC_A | 3.73e-77 | 110 | 516 | 14 | 393 | GH5endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYC_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYD_A GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYD_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYE_A GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYE_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107] |
| 6HA9_A | 2.88e-76 | 110 | 516 | 14 | 393 | Structureof an endo-Xyloglucanase from Cellvibrio japonicus complexed with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107],6HA9_B Structure of an endo-Xyloglucanase from Cellvibrio japonicus complexed with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107],6HAA_A Structure of a covalent complex of endo-Xyloglucanase from Cellvibrio japonicus after reacting with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107],6HAA_B Structure of a covalent complex of endo-Xyloglucanase from Cellvibrio japonicus after reacting with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107] |
| 1EDG_A | 6.69e-68 | 130 | 474 | 12 | 345 | SingleCrystal Structure Determination Of The Catalytic Domain Of Celcca Carried Out At 15 Degree C [Ruminiclostridium cellulolyticum H10] |
| 3ZMR_A | 1.28e-63 | 88 | 480 | 60 | 447 | Bacteroidesovatus GH5 xyloglucanase in complex with a XXXG heptasaccharide [Bacteroides ovatus],3ZMR_B Bacteroides ovatus GH5 xyloglucanase in complex with a XXXG heptasaccharide [Bacteroides ovatus] |
| 6XSU_A | 4.79e-63 | 145 | 514 | 13 | 351 | GH5-4broad specificity endoglucanase from Ruminococcus flavefaciens [Ruminococcus flavefaciens],6XSU_B GH5-4 broad specificity endoglucanase from Ruminococcus flavefaciens [Ruminococcus flavefaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P17901 | 4.37e-66 | 130 | 474 | 37 | 370 | Endoglucanase A OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCA PE=1 SV=1 |
| A7LXT7 | 1.32e-62 | 88 | 480 | 95 | 482 | Xyloglucan-specific endo-beta-1,4-glucanase BoGH5A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02653 PE=1 SV=1 |
| O08342 | 1.59e-62 | 145 | 517 | 40 | 400 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| P23660 | 1.49e-58 | 145 | 517 | 27 | 364 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
| P54937 | 1.07e-57 | 118 | 521 | 12 | 383 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
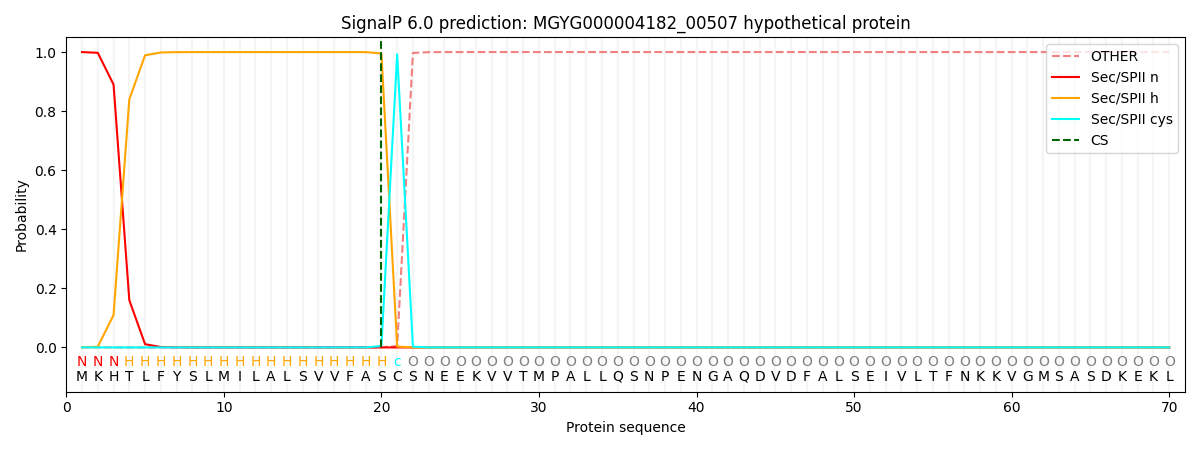
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000067 | 0.000000 | 0.000000 | 0.000000 |
