You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004197_00807
You are here: Home > Sequence: MGYG000004197_00807
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes_A; | |||||||||||
| CAZyme ID | MGYG000004197_00807 | |||||||||||
| CAZy Family | GH127 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2533; End: 4611 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH127 | 115 | 573 | 6.6e-64 | 0.898854961832061 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07944 | Glyco_hydro_127 | 1.11e-68 | 113 | 574 | 53 | 503 | Beta-L-arabinofuranosidase, GH127. One member of this family, from Bidobacterium longicum, UniProtKB:E8MGH8, has been characterized as an unusual beta-L-arabinofuranosidase enzyme, EC:3.2.1.185. It rleases l-arabinose from the l-arabinofuranose (Araf)-beta1,2-Araf disaccharide and also transglycosylates 1-alkanols with retention of the anomeric configuration. Terminal beta-l-arabinofuranosyl residues have been found in arabinogalactan proteins from a mumber of different plantt species. beta-l-Arabinofuranosyl linkages with 1-4 arabinofuranosides are also found in the sugar chains of extensin and solanaceous lectins, hydroxyproline (Hyp)2-rich glycoproteins that are widely observed in plant cell wall fractions. The critical residue for catalytic activity is Glu-338, in a ET/SCAS sequence context. |
| COG3533 | COG3533 | 2.69e-60 | 60 | 576 | 2 | 503 | Uncharacterized conserved protein, DUF1680 family [Function unknown]. |
| cd04791 | LanC_SerThrkinase | 2.81e-06 | 182 | 260 | 93 | 173 | Lanthionine synthetase C-like domain associated with serine/threonine kinases. Some members of this subgroup lack the zinc binding site and the active site residues, and therefore are most likely inactive. The function of this domain is unknown. |
| COG4225 | YesR | 6.10e-04 | 170 | 272 | 32 | 121 | Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism]. |
| cd04792 | LanM-like | 0.001 | 182 | 260 | 599 | 681 | Cyclases involved in the biosynthesis of class II lantibiotics, and similar proteins. LanM-like proteins. LanM is a bifunctional enzyme, involved in the synthesis of class II lantibiotics. It is responsible for both the dehydration and the cyclization of the precursor-peptide during lantibiotic synthesis. The C-terminal domain shows similarity to LanC, the cyclase component of the lan operon, but the N terminus seems to be unrelated to the dehydratase, LanB. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARE60508.1 | 3.47e-118 | 79 | 656 | 31 | 605 |
| QWG10444.1 | 1.00e-45 | 79 | 606 | 47 | 614 |
| AZQ63834.1 | 3.97e-44 | 79 | 598 | 47 | 610 |
| QHS51661.1 | 1.05e-43 | 122 | 576 | 123 | 582 |
| CAB9494536.1 | 2.96e-43 | 66 | 588 | 21 | 581 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQO_A | 1.35e-31 | 31 | 577 | 5 | 610 | Glycosidehydrolase BT_1003 [Bacteroides thetaiotaomicron] |
| 6EX6_A | 1.01e-26 | 128 | 572 | 76 | 543 | TheGH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482],6EX6_B The GH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482] |
| 3WRE_A | 2.36e-25 | 122 | 572 | 80 | 564 | Thecrystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 [Bifidobacterium longum subsp. longum JCM 1217],3WRG_A The complex structure of HypBA1 with L-arabinose [Bifidobacterium longum subsp. longum JCM 1217] |
| 3WKW_A | 2.44e-25 | 122 | 572 | 80 | 564 | Crystalstructure of GH127 beta-L-arabinofuranosidase HypBA1 from Bifidobacterium longum ligand free form [Bifidobacterium longum subsp. longum JCM 1217],3WKX_A Crystal structure of GH127 beta-L-arabinofuranosidase HypBA1 from Bifidobacterium longum arabinose complex form [Bifidobacterium longum subsp. longum JCM 1217],7BZL_A Chain A, Non-reducing end beta-L-arabinofuranosidase [Bifidobacterium longum subsp. longum JCM 1217],7DIF_A Chain A, Non-reducing end beta-L-arabinofuranosidase [Bifidobacterium longum subsp. longum JCM 1217],7EXV_A Chain A, Non-reducing end beta-L-arabinofuranosidase [Bifidobacterium longum subsp. longum JCM 1217],7EXW_A Chain A, Non-reducing end beta-L-arabinofuranosidase [Bifidobacterium longum subsp. longum JCM 1217] |
| 7EXU_A | 5.70e-25 | 122 | 572 | 80 | 564 | ChainA, Non-reducing end beta-L-arabinofuranosidase [Bifidobacterium longum subsp. longum JCM 1217] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH8 | 1.29e-24 | 122 | 572 | 80 | 564 | Non-reducing end beta-L-arabinofuranosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
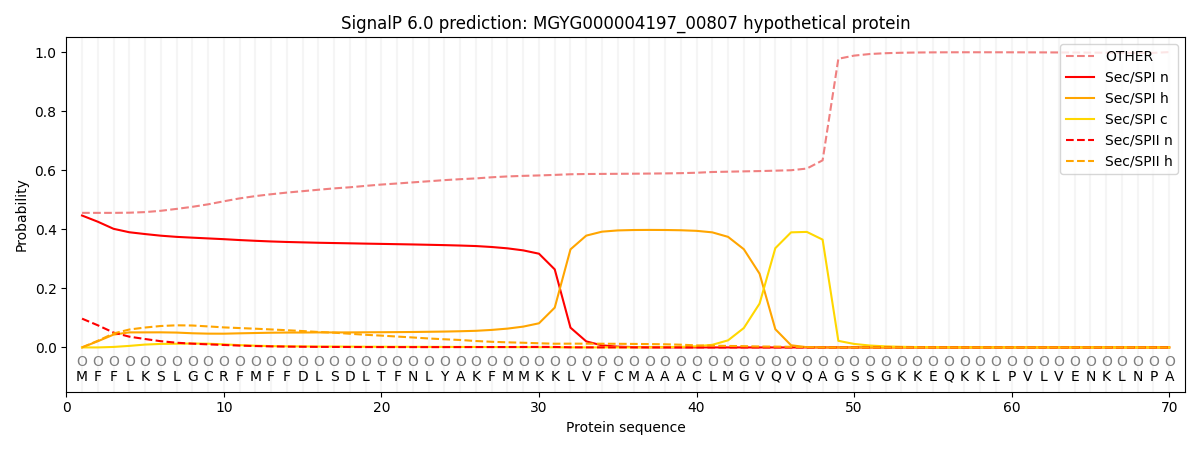
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.465427 | 0.429802 | 0.103276 | 0.000544 | 0.000432 | 0.000516 |
