You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004203_01379
You are here: Home > Sequence: MGYG000004203_01379
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
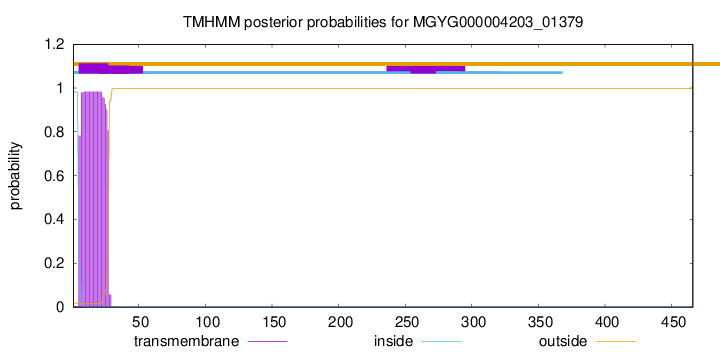
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Acetatifactor; | |||||||||||
| CAZyme ID | MGYG000004203_01379 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 65466; End: 66866 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 164 | 461 | 2.3e-64 | 0.9867986798679867 |
| CBM32 | 41 | 156 | 2.3e-27 | 0.9354838709677419 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 9.48e-29 | 164 | 462 | 3 | 311 | Glycosyl hydrolase family 26. |
| pfam00754 | F5_F8_type_C | 5.10e-23 | 43 | 154 | 4 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 7.44e-14 | 43 | 159 | 16 | 143 | Substituted updates: Jan 31, 2002 |
| COG4124 | ManB2 | 6.37e-11 | 259 | 397 | 119 | 266 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| smart00231 | FA58C | 1.75e-08 | 44 | 159 | 15 | 138 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGG55380.1 | 1.99e-136 | 13 | 464 | 46 | 504 |
| QJC51244.1 | 9.83e-135 | 34 | 464 | 41 | 474 |
| ASS68840.1 | 8.81e-129 | 30 | 464 | 165 | 603 |
| ATB27078.1 | 8.20e-122 | 170 | 464 | 199 | 493 |
| ATL47771.1 | 1.62e-112 | 160 | 464 | 30 | 339 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q75_A | 6.11e-55 | 165 | 463 | 27 | 327 | Thestructure of GH26A from Muricauda sp. MAR_2010_75 [Muricauda sp. MAR_2010_75],6Q75_B The structure of GH26A from Muricauda sp. MAR_2010_75 [Muricauda sp. MAR_2010_75] |
| 2RV9_A | 3.97e-30 | 34 | 159 | 7 | 136 | Solutionstructure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 4ZXE_A | 4.09e-30 | 34 | 159 | 8 | 137 | X-raycrystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_B X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_C X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis] |
| 4ZY9_A | 2.09e-29 | 34 | 159 | 8 | 137 | X-raycrystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZY9_B X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 2RVA_A | 6.84e-26 | 34 | 159 | 7 | 137 | Solutionstructure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P55278 | 6.81e-22 | 201 | 456 | 73 | 345 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis OX=1423 PE=3 SV=1 |
| P16699 | 7.96e-21 | 208 | 390 | 84 | 273 | Mannan endo-1,4-beta-mannosidase A and B OS=Caldalkalibacillus mannanilyticus (strain DSM 16130 / CIP 109019 / JCM 10596 / AM-001) OX=1236954 PE=1 SV=1 |
| Q5AWB7 | 1.82e-20 | 295 | 435 | 181 | 319 | Probable mannan endo-1,4-beta-mannosidase E OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manE PE=3 SV=1 |
| G2Q4H7 | 2.31e-19 | 160 | 463 | 175 | 476 | Mannan endo-1,4-beta-mannosidase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Man26A PE=1 SV=1 |
| O05512 | 3.00e-19 | 201 | 398 | 75 | 279 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis (strain 168) OX=224308 GN=gmuG PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
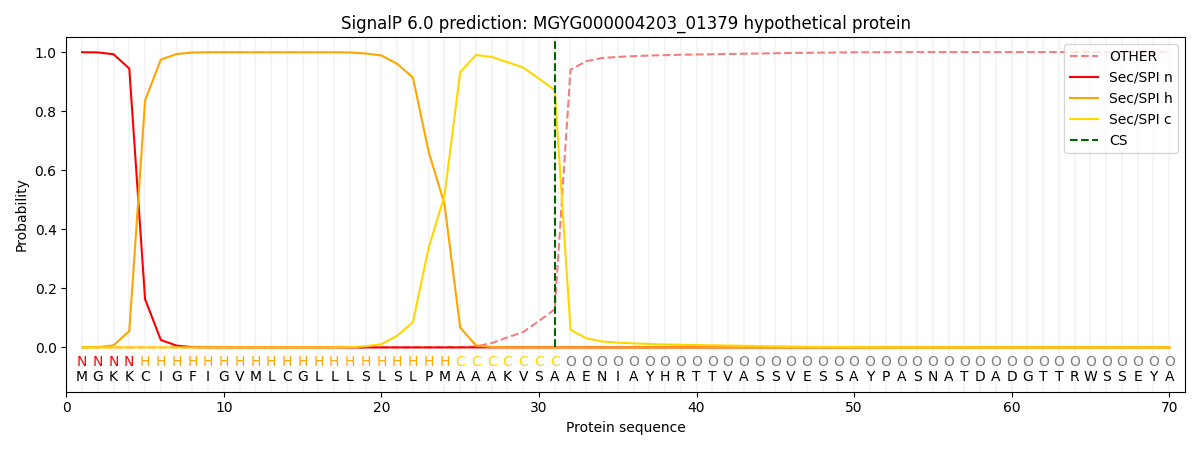
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001188 | 0.997671 | 0.000476 | 0.000271 | 0.000201 | 0.000176 |