You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004203_01380
You are here: Home > Sequence: MGYG000004203_01380
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
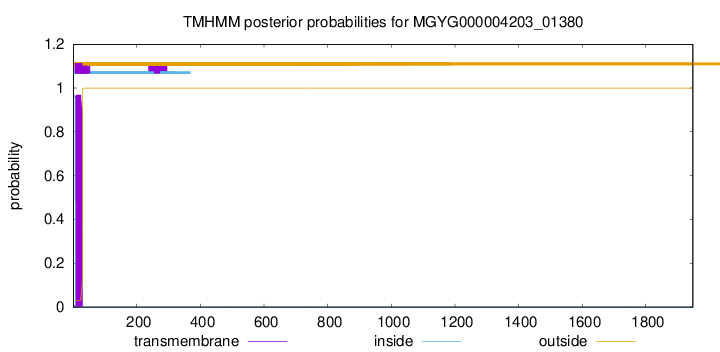
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Acetatifactor; | |||||||||||
| CAZyme ID | MGYG000004203_01380 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 66891; End: 72737 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 109 | 376 | 1.5e-70 | 0.9765625 |
| CBM23 | 925 | 1082 | 1.4e-33 | 0.9691358024691358 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd00118 | LysM | 1.02e-14 | 1845 | 1890 | 1 | 45 | Lysin Motif is a small domain involved in binding peptidoglycan. LysM, a small globular domain with approximately 40 amino acids, is a widespread protein module involved in binding peptidoglycan in bacteria and chitin in eukaryotes. The domain was originally identified in enzymes that degrade bacterial cell walls, but proteins involved in many other biological functions also contain this domain. It has been reported that the LysM domain functions as a signal for specific plant-bacteria recognition in bacterial pathogenesis. Many of these enzymes are modular and are composed of catalytic units linked to one or several repeats of LysM domains. LysM domains are found in bacteria and eukaryotes. |
| PRK06347 | PRK06347 | 5.82e-14 | 1813 | 1947 | 306 | 449 | 1,4-beta-N-acetylmuramoylhydrolase. |
| smart00257 | LysM | 2.67e-13 | 1847 | 1890 | 2 | 44 | Lysin motif. |
| TIGR02899 | spore_safA | 3.21e-13 | 1849 | 1890 | 1 | 42 | spore coat assembly protein SafA. SafA (YrbB) (SafA) of Bacillus subtilis is a protein found at the interface of the spore cortex and spore coat, and is dependent on SpoVID for its localization. This model is based on the N-terminal LysM (lysin motif) domain (see pfamAM model pfam01476) of SafA and, from several other spore-forming species, the protein with the most similar N-terminal region. However, this set of proteins differs greatly in C-terminal of the LysM domaim; blocks of 12-residue and 13-residue repeats are found in the Bacillus cereus group, tandem LysM domains in Thermoanaerobacter tengcongensis MB4, etc. in which one of which is found in most examples of endospore-forming bacteria. [Cellular processes, Sporulation and germination] |
| pfam01476 | LysM | 6.22e-12 | 1847 | 1890 | 1 | 42 | LysM domain. The LysM (lysin motif) domain is about 40 residues long. It is found in a variety of enzymes involved in bacterial cell wall degradation. This domain may have a general peptidoglycan binding function. The structure of this domain is known. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHW31794.1 | 1.20e-208 | 60 | 1483 | 37 | 1546 |
| QJC50841.1 | 6.19e-196 | 69 | 1354 | 57 | 1420 |
| QGG54903.1 | 2.73e-195 | 69 | 1329 | 60 | 1402 |
| ASS68430.2 | 1.50e-193 | 62 | 1348 | 52 | 1418 |
| AYQ73430.1 | 6.29e-178 | 61 | 1100 | 47 | 1130 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4QP0_A | 2.11e-18 | 74 | 348 | 4 | 300 | CrystalStructure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
| 1UUQ_A | 8.80e-18 | 72 | 292 | 22 | 254 | Exo-mannosidasefrom Cellvibrio mixtus [Cellvibrio mixtus],1UZ4_A Common inhibition of beta-glucosidase and beta-mannosidase by isofagomine lactam reflects different conformational intineraries for glucoside and mannoside hydrolysis [Cellvibrio mixtus],7ODJ_AAA Chain AAA, Man5A [Cellvibrio mixtus] |
| 3PZ9_A | 1.22e-17 | 153 | 292 | 96 | 223 | Nativestructure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
| 6TN6_A | 5.12e-17 | 153 | 292 | 82 | 209 | X-raystructure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
| 1RH9_A | 6.50e-15 | 73 | 289 | 5 | 210 | ChainA, endo-beta-mannanase [Solanum lycopersicum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O48540 | 2.20e-20 | 151 | 374 | 100 | 359 | Mannan endo-1,4-beta-mannosidase 1 OS=Solanum lycopersicum OX=4081 GN=MAN1 PE=1 SV=2 |
| Q0JKM9 | 2.54e-18 | 151 | 289 | 111 | 242 | Mannan endo-1,4-beta-mannosidase 1 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN1 PE=2 SV=2 |
| Q9SG95 | 2.05e-16 | 150 | 292 | 102 | 237 | Putative mannan endo-1,4-beta-mannosidase 4 OS=Arabidopsis thaliana OX=3702 GN=MAN4 PE=3 SV=1 |
| Q9FUQ6 | 3.41e-16 | 151 | 374 | 101 | 361 | Mannan endo-1,4-beta-mannosidase 3 OS=Solanum lycopersicum OX=4081 GN=MAN3 PE=3 SV=1 |
| Q7Y223 | 7.98e-16 | 151 | 292 | 115 | 250 | Mannan endo-1,4-beta-mannosidase 2 OS=Arabidopsis thaliana OX=3702 GN=MAN2 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
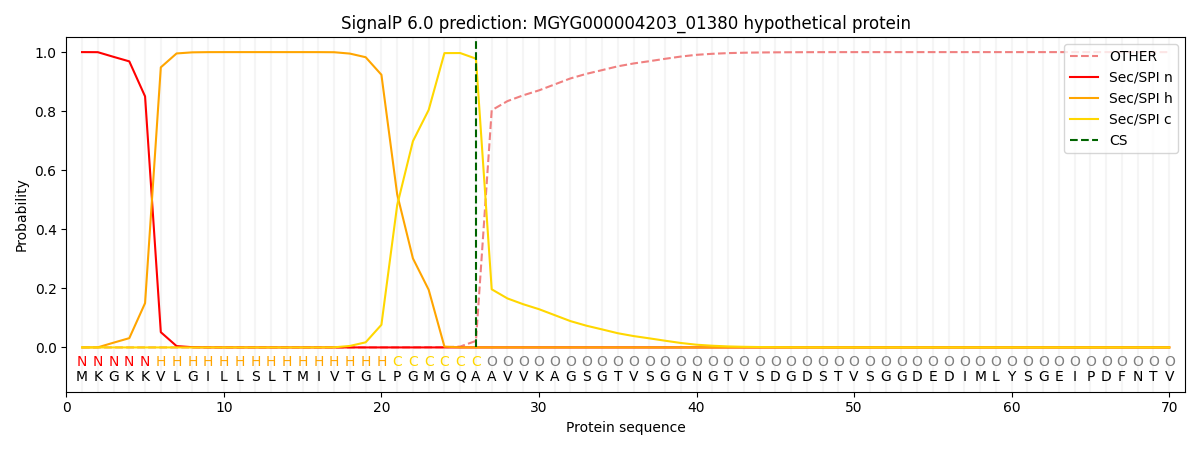
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000296 | 0.998923 | 0.000251 | 0.000187 | 0.000166 | 0.000144 |