You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004221_01279
You are here: Home > Sequence: MGYG000004221_01279
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
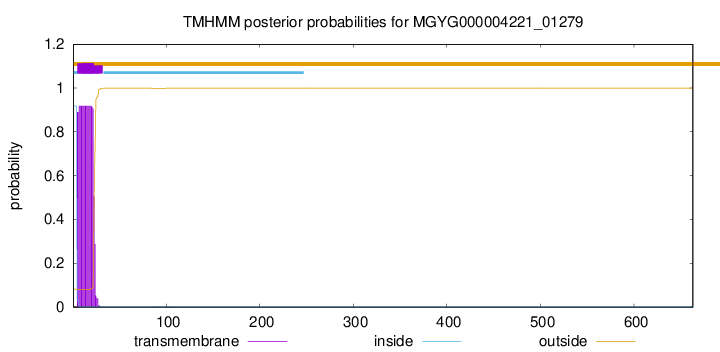
TMHMM annotations
Basic Information help
| Species | UMGS621 sp900543915 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; Borkfalkiaceae; UMGS621; UMGS621 sp900543915 | |||||||||||
| CAZyme ID | MGYG000004221_01279 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26713; End: 28704 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 371 | 642 | 3.2e-40 | 0.796923076923077 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 5.43e-16 | 417 | 580 | 237 | 402 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| PLN03003 | PLN03003 | 2.14e-04 | 417 | 566 | 137 | 291 | Probable polygalacturonase At3g15720 |
| pfam00295 | Glyco_hydro_28 | 3.80e-04 | 420 | 648 | 87 | 315 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PLN02155 | PLN02155 | 0.007 | 420 | 489 | 147 | 218 | polygalacturonase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNK57811.1 | 1.34e-118 | 1 | 651 | 1 | 640 |
| BBH22092.1 | 2.50e-117 | 1 | 651 | 4 | 647 |
| QTH44486.1 | 9.32e-110 | 1 | 649 | 1 | 645 |
| QJD87459.1 | 2.46e-108 | 14 | 649 | 22 | 643 |
| AMB60207.1 | 3.09e-97 | 100 | 651 | 1 | 556 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3JUR_A | 2.30e-07 | 417 | 534 | 189 | 316 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
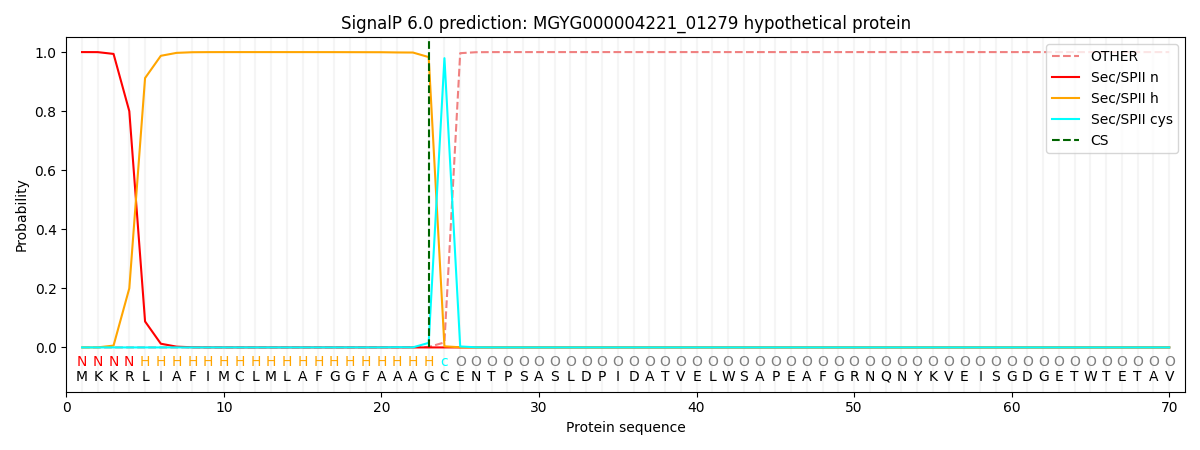
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000010 | 1.000045 | 0.000000 | 0.000000 | 0.000000 |