You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004223_01295
You are here: Home > Sequence: MGYG000004223_01295
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS1883 sp900763305 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; UMGS1883; UMGS1883; UMGS1883; UMGS1883 sp900763305 | |||||||||||
| CAZyme ID | MGYG000004223_01295 | |||||||||||
| CAZy Family | PL35 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 57280; End: 61671 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL35 | 998 | 1156 | 7.6e-34 | 0.9050279329608939 |
| CBM32 | 1341 | 1456 | 2.6e-21 | 0.9193548387096774 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 3.05e-15 | 1343 | 1458 | 4 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam16332 | DUF4962 | 3.80e-08 | 699 | 797 | 209 | 303 | Domain of unknown function (DUF4962). This family consists of uncharacterized proteins around 870 residues in length and is mainly found in various Bacteroides species. The function of this protein is unknown. |
| cd00057 | FA58C | 7.95e-04 | 1327 | 1442 | 4 | 126 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUH30172.1 | 6.55e-107 | 636 | 1460 | 56 | 892 |
| ALS27107.1 | 3.19e-104 | 606 | 1460 | 654 | 1510 |
| AZS17848.1 | 8.03e-97 | 361 | 1460 | 166 | 1276 |
| QNK58643.1 | 4.12e-90 | 631 | 1460 | 772 | 1608 |
| AYV68237.1 | 2.17e-85 | 635 | 1202 | 53 | 626 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7D29_A | 3.13e-23 | 1345 | 1460 | 16 | 131 | CBM32of AlyQ [Persicobacter sp. CCB-QB2],7D2A_A CBM32 of AlyQ in complex with 4,5-unsaturated mannuronic acid [Persicobacter sp. CCB-QB2] |
| 5XNR_A | 5.17e-21 | 1345 | 1460 | 16 | 131 | TruncatedAlyQ with CBM32 and alginate lyase domains [Persicobacter sp. CCB-QB2] |
| 5ZU6_A | 1.15e-16 | 1322 | 1456 | 23 | 152 | ACBM32 derived from alginate lyase B (AlyB-OU02) [Vibrio] |
| 5ZU5_A | 8.27e-15 | 1322 | 1456 | 23 | 152 | Crystalstructure of a full length alginate lyase with CBM domain [Vibrio splendidus] |
| 2JD9_A | 1.03e-09 | 1348 | 1460 | 21 | 133 | Structureof a pectin binding carbohydrate binding module determined in an orthorhombic crystal form. [Yersinia enterocolitica],2JDA_A Structure of a pectin binding carbohydrate binding module determined in an monoclinic crystal form. [Yersinia enterocolitica],2JDA_B Structure of a pectin binding carbohydrate binding module determined in an monoclinic crystal form. [Yersinia enterocolitica] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
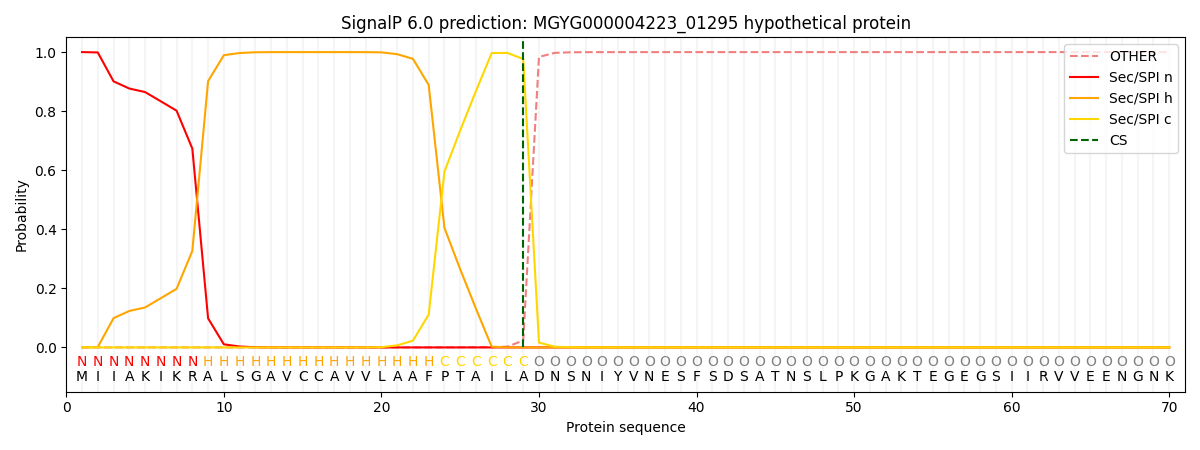
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000296 | 0.998976 | 0.000252 | 0.000164 | 0.000155 | 0.000146 |
