You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004223_01592
You are here: Home > Sequence: MGYG000004223_01592
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS1883 sp900763305 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; UMGS1883; UMGS1883; UMGS1883; UMGS1883 sp900763305 | |||||||||||
| CAZyme ID | MGYG000004223_01592 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 42847; End: 44505 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 348 | 490 | 2.7e-29 | 0.40594059405940597 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4124 | ManB2 | 5.02e-18 | 236 | 514 | 34 | 320 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| pfam02156 | Glyco_hydro_26 | 7.49e-14 | 346 | 458 | 126 | 243 | Glycosyl hydrolase family 26. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOX62356.1 | 8.14e-135 | 65 | 552 | 38 | 511 |
| QUH19438.1 | 4.67e-124 | 74 | 551 | 59 | 523 |
| QIB68979.1 | 8.36e-122 | 74 | 552 | 44 | 506 |
| QHI71412.1 | 1.48e-119 | 74 | 551 | 44 | 507 |
| QAT42469.1 | 7.22e-118 | 74 | 551 | 47 | 510 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VI0_A | 1.14e-24 | 343 | 549 | 78 | 278 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2BVD_A | 2.15e-24 | 343 | 522 | 78 | 257 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
| 2V3G_A | 2.15e-24 | 343 | 522 | 78 | 257 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2BV9_A | 2.45e-24 | 343 | 522 | 78 | 257 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2CIP_A | 5.30e-24 | 343 | 522 | 78 | 257 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16218 | 4.39e-22 | 343 | 522 | 100 | 279 | Endoglucanase H OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celH PE=1 SV=1 |
| P55278 | 1.38e-07 | 346 | 456 | 160 | 271 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis OX=1423 PE=3 SV=1 |
| Q5AWB7 | 7.43e-07 | 353 | 451 | 181 | 265 | Probable mannan endo-1,4-beta-mannosidase E OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
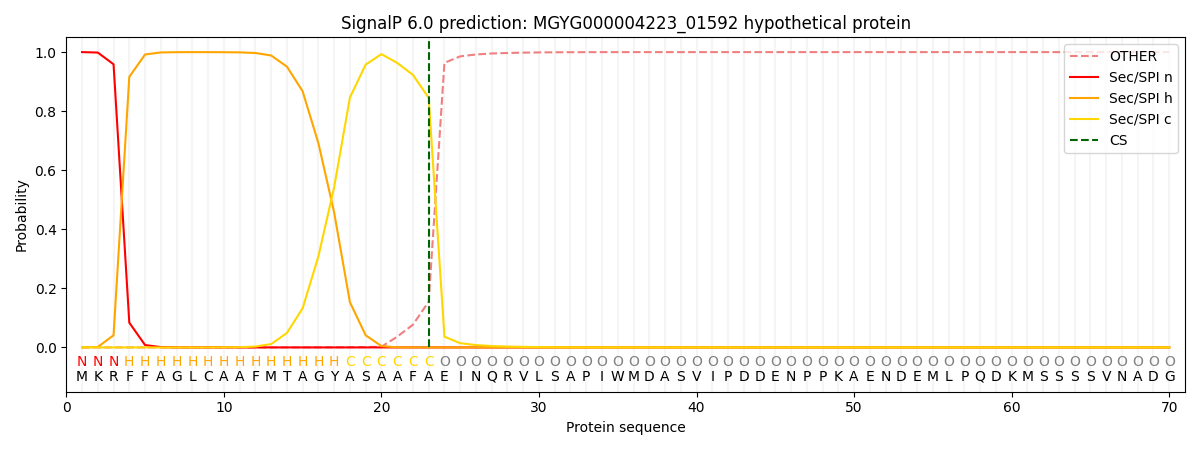
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001000 | 0.998067 | 0.000271 | 0.000250 | 0.000210 | 0.000186 |
