You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004230_00155
You are here: Home > Sequence: MGYG000004230_00155
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium sp900759995 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium sp900759995 | |||||||||||
| CAZyme ID | MGYG000004230_00155 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 40820; End: 43720 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 53 | 199 | 2.2e-27 | 0.9850746268656716 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 6.37e-30 | 51 | 199 | 2 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 1.25e-19 | 53 | 199 | 6 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam03174 | CHB_HEX_C | 7.51e-04 | 805 | 836 | 37 | 67 | Chitobiase/beta-hexosaminidase C-terminal domain. This short domain represents the C terminal domain in chitobiases and beta-hexosaminidases EC:3.2.1.52. It is composed of a beta sandwich structure. The function of this domain is unknown. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCL58565.1 | 2.17e-246 | 48 | 856 | 77 | 904 |
| AMN35280.1 | 4.44e-181 | 23 | 854 | 21 | 842 |
| AQW23377.1 | 4.84e-180 | 9 | 854 | 6 | 842 |
| ATD49073.1 | 5.56e-180 | 9 | 854 | 11 | 847 |
| QGM19047.1 | 3.55e-176 | 49 | 854 | 137 | 948 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JS4_A | 2.29e-15 | 53 | 232 | 613 | 799 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 2VMG_A | 5.35e-13 | 41 | 199 | 4 | 156 | Thestructure of CBM51 from Clostridium perfringens GH95 in complex with methyl-galactose [Clostridium perfringens] |
| 2VMH_A | 8.59e-13 | 52 | 199 | 9 | 150 | Thestructure of CBM51 from Clostridium perfringens GH95 [Clostridium perfringens] |
| 2VMI_A | 1.01e-11 | 52 | 199 | 9 | 150 | Thestructure of seleno-methionine labelled CBM51 from Clostridium perfringens GH95 [Clostridium perfringens] |
| 7JRM_A | 1.86e-08 | 40 | 263 | 79 | 285 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
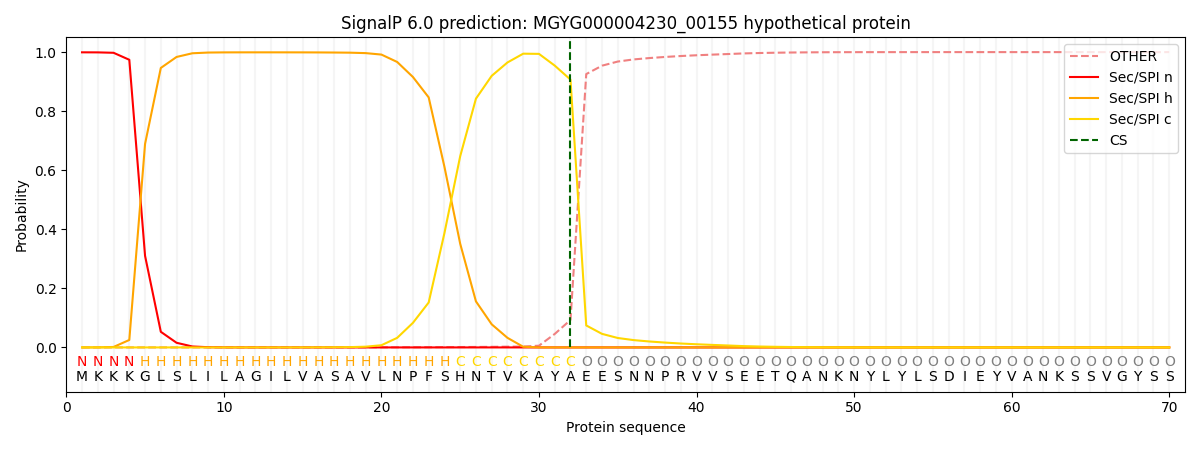
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000508 | 0.997774 | 0.000991 | 0.000310 | 0.000205 | 0.000169 |
