You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004230_01242
You are here: Home > Sequence: MGYG000004230_01242
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
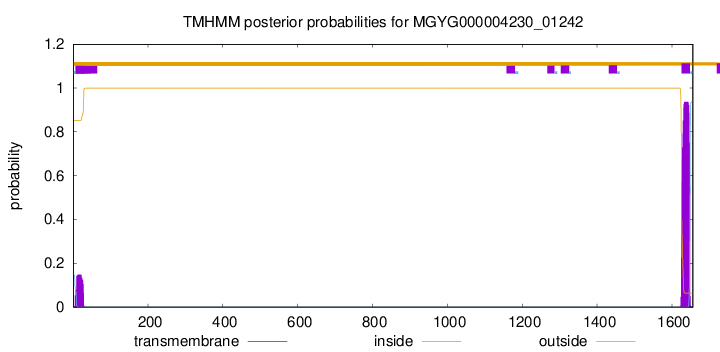
TMHMM annotations
Basic Information help
| Species | Clostridium sp900759995 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium sp900759995 | |||||||||||
| CAZyme ID | MGYG000004230_01242 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 27350; End: 32320 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 1233 | 1356 | 1.5e-18 | 0.9112903225806451 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 5.32e-17 | 1226 | 1355 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 2.95e-08 | 136 | 247 | 16 | 110 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 2.85e-06 | 136 | 252 | 27 | 131 | Substituted updates: Jan 31, 2002 |
| COG0419 | SbcC | 1.70e-05 | 1358 | 1573 | 210 | 432 | DNA repair exonuclease SbcCD ATPase subunit [Replication, recombination and repair]. |
| TIGR01167 | LPXTG_anchor | 6.59e-05 | 1621 | 1654 | 1 | 34 | LPXTG-motif cell wall anchor domain. This model describes the LPXTG motif-containing region found at the C-terminus of many surface proteins of Streptococcus and Streptomyces species. Cleavage between the Thr and Gly by sortase or a related enzyme leads to covalent anchoring at the new C-terminal Thr to the cell wall. Hits that do not lie at the C-terminus or are not found in Gram-positive bacteria are probably false-positive. A common feature of this proteins containing this domain appears to be a high proportion of charged and zwitterionic residues immediatedly upstream of the LPXTG motif. This model differs from other descriptions of the LPXTG region by including a portion of that upstream charged region. [Cell envelope, Other] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABG83191.1 | 0.0 | 1 | 1488 | 1 | 1490 |
| SQG39183.1 | 0.0 | 1 | 1488 | 1 | 1490 |
| AQW23840.1 | 0.0 | 1 | 1488 | 1 | 1490 |
| AOY54034.1 | 0.0 | 1 | 1488 | 1 | 1490 |
| AXH52486.1 | 0.0 | 1 | 1488 | 1 | 1490 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2V72_A | 4.34e-12 | 1226 | 1359 | 9 | 143 | Thestructure of the family 32 CBM from C. perfringens NanJ in complex with galactose [Clostridium perfringens] |
| 1W8N_A | 6.10e-06 | 1239 | 1357 | 477 | 597 | Contributionof the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. [Micromonospora viridifaciens],1W8O_A Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens [Micromonospora viridifaciens] |
| 1WCQ_A | 6.10e-06 | 1239 | 1357 | 477 | 597 | Mutagenesisof the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_B Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_C Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens] |
| 2BER_A | 6.10e-06 | 1239 | 1357 | 477 | 597 | Y370GActive Site Mutant of the Sialidase from Micromonospora viridifaciens in complex with beta-Neu5Ac (sialic acid). [Micromonospora viridifaciens] |
| 2BZD_A | 6.10e-06 | 1239 | 1357 | 477 | 597 | Galactoserecognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_B Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_C Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
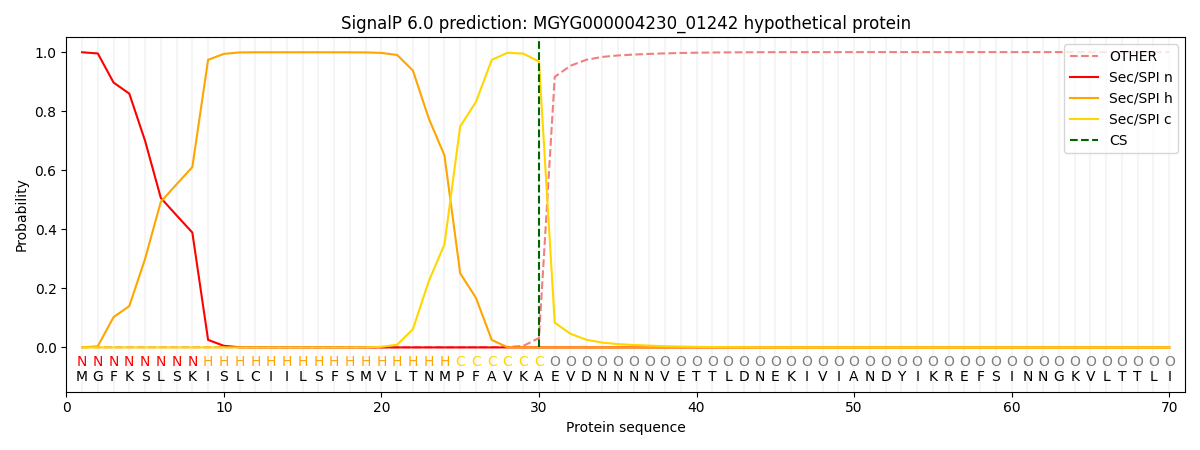
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001727 | 0.997273 | 0.000262 | 0.000270 | 0.000225 | 0.000204 |