You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004230_01456
You are here: Home > Sequence: MGYG000004230_01456
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
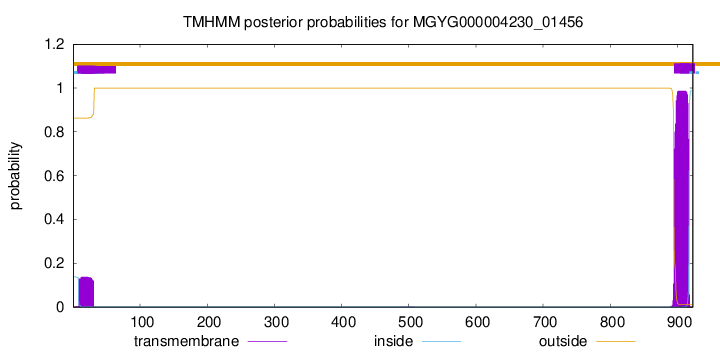
TMHMM annotations
Basic Information help
| Species | Clostridium sp900759995 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium sp900759995 | |||||||||||
| CAZyme ID | MGYG000004230_01456 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3125; End: 5896 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 325 | 683 | 6e-48 | 0.9385964912280702 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 1.90e-59 | 324 | 690 | 6 | 339 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| cd00063 | FN3 | 6.89e-11 | 741 | 824 | 1 | 92 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| smart00060 | FN3 | 2.56e-08 | 741 | 816 | 1 | 82 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
| pfam00041 | fn3 | 3.17e-07 | 744 | 815 | 3 | 80 | Fibronectin type III domain. |
| TIGR01167 | LPXTG_anchor | 2.38e-04 | 889 | 923 | 2 | 34 | LPXTG-motif cell wall anchor domain. This model describes the LPXTG motif-containing region found at the C-terminus of many surface proteins of Streptococcus and Streptomyces species. Cleavage between the Thr and Gly by sortase or a related enzyme leads to covalent anchoring at the new C-terminal Thr to the cell wall. Hits that do not lie at the C-terminus or are not found in Gram-positive bacteria are probably false-positive. A common feature of this proteins containing this domain appears to be a high proportion of charged and zwitterionic residues immediatedly upstream of the LPXTG motif. This model differs from other descriptions of the LPXTG region by including a portion of that upstream charged region. [Cell envelope, Other] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWG19756.1 | 1.15e-172 | 315 | 775 | 115 | 576 |
| AVO06163.1 | 1.15e-172 | 315 | 775 | 115 | 576 |
| AWI02704.1 | 1.15e-172 | 315 | 775 | 115 | 576 |
| AVO03208.1 | 1.15e-172 | 315 | 775 | 115 | 576 |
| AAS10966.1 | 1.93e-103 | 294 | 735 | 105 | 541 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4FJ6_A | 3.37e-20 | 297 | 674 | 149 | 504 | Crystalstructure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution [Parabacteroides distasonis ATCC 8503],4FJ6_B Crystal structure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution [Parabacteroides distasonis ATCC 8503],4FJ6_C Crystal structure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution [Parabacteroides distasonis ATCC 8503],4FJ6_D Crystal structure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution [Parabacteroides distasonis ATCC 8503] |
| 6MYV_A | 1.02e-18 | 327 | 674 | 176 | 504 | Sialidase26co-crystallized with DANA-Gc [bacterium],6MYV_B Sialidase26 co-crystallized with DANA-Gc [bacterium],6MYV_C Sialidase26 co-crystallized with DANA-Gc [bacterium],6MYV_D Sialidase26 co-crystallized with DANA-Gc [bacterium] |
| 6MRV_A | 1.14e-18 | 327 | 674 | 197 | 525 | Sialidase26co-crystallized with DANA [bacterium],6MRV_B Sialidase26 co-crystallized with DANA [bacterium],6MRX_A Sialidase26 apo [bacterium],6MRX_B Sialidase26 apo [bacterium],6MRX_C Sialidase26 apo [bacterium],6MRX_D Sialidase26 apo [bacterium] |
| 4Q6K_A | 6.50e-16 | 327 | 674 | 179 | 507 | Crystalstructure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target) [Bacteroides caccae ATCC 43185],4Q6K_B Crystal structure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target) [Bacteroides caccae ATCC 43185],4Q6K_C Crystal structure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target) [Bacteroides caccae ATCC 43185],4Q6K_D Crystal structure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target) [Bacteroides caccae ATCC 43185] |
| 6MNJ_A | 1.84e-13 | 332 | 674 | 200 | 521 | Hadzamicrobial sialidase Hz136 [Alistipes],6MNJ_B Hadza microbial sialidase Hz136 [Alistipes] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P15698 | 1.00e-17 | 315 | 674 | 35 | 380 | Sialidase OS=Paeniclostridium sordellii OX=1505 PE=1 SV=1 |
| P31206 | 1.24e-15 | 332 | 674 | 203 | 526 | Sialidase OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=nanH PE=3 SV=2 |
| P29767 | 8.51e-10 | 328 | 676 | 391 | 811 | Sialidase OS=Clostridium septicum OX=1504 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
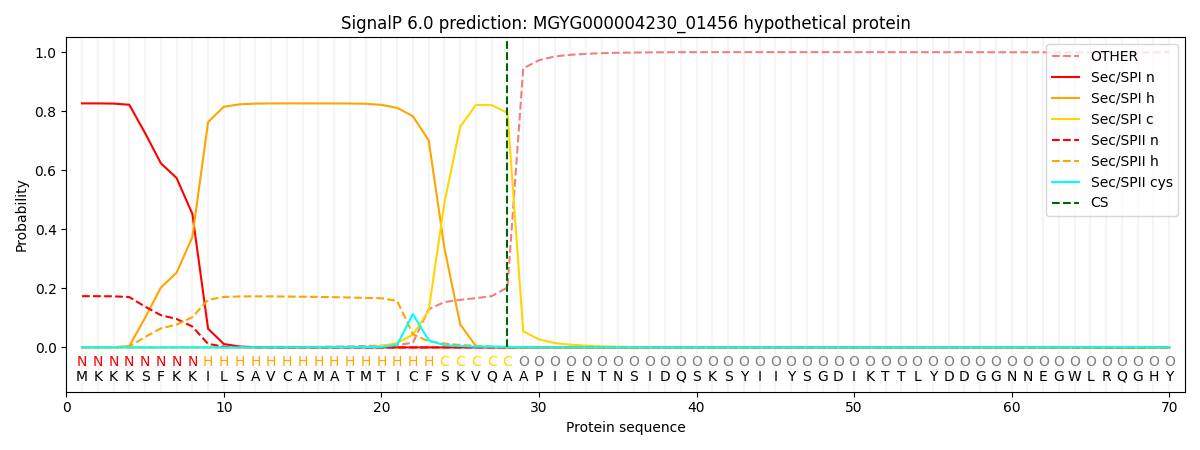
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000514 | 0.817621 | 0.180996 | 0.000317 | 0.000290 | 0.000233 |