You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004230_01906
You are here: Home > Sequence: MGYG000004230_01906
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
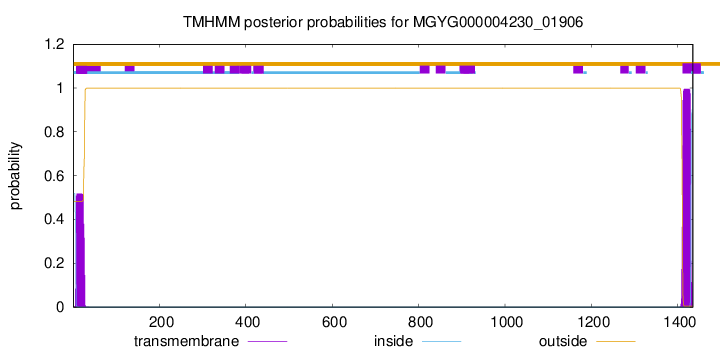
TMHMM annotations
Basic Information help
| Species | Clostridium sp900759995 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium sp900759995 | |||||||||||
| CAZyme ID | MGYG000004230_01906 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8522; End: 12829 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 67 | 213 | 2.9e-25 | 0.9925373134328358 |
| CBM32 | 891 | 1009 | 9.1e-18 | 0.9193548387096774 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 8.23e-28 | 66 | 214 | 2 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 1.88e-15 | 63 | 213 | 1 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam00754 | F5_F8_type_C | 8.23e-14 | 892 | 1008 | 4 | 120 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 1.80e-05 | 892 | 1008 | 17 | 136 | Substituted updates: Jan 31, 2002 |
| PRK08581 | PRK08581 | 0.001 | 1325 | 1406 | 37 | 118 | amidase domain-containing protein. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCL58565.1 | 1.75e-315 | 68 | 1165 | 82 | 1191 |
| SQG16429.1 | 1.60e-291 | 63 | 1086 | 139 | 1158 |
| VED84782.1 | 1.60e-291 | 63 | 1086 | 139 | 1158 |
| QTZ58125.1 | 5.94e-291 | 63 | 1086 | 137 | 1156 |
| VTP91146.1 | 6.32e-291 | 63 | 1086 | 139 | 1158 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VMH_A | 3.15e-16 | 62 | 213 | 4 | 150 | Thestructure of CBM51 from Clostridium perfringens GH95 [Clostridium perfringens] |
| 2VMG_A | 3.68e-16 | 62 | 213 | 10 | 156 | Thestructure of CBM51 from Clostridium perfringens GH95 in complex with methyl-galactose [Clostridium perfringens] |
| 2VMI_A | 1.09e-15 | 62 | 213 | 4 | 150 | Thestructure of seleno-methionine labelled CBM51 from Clostridium perfringens GH95 [Clostridium perfringens] |
| 7JRM_A | 4.71e-10 | 69 | 252 | 86 | 247 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JRL_A | 5.00e-10 | 69 | 252 | 108 | 269 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
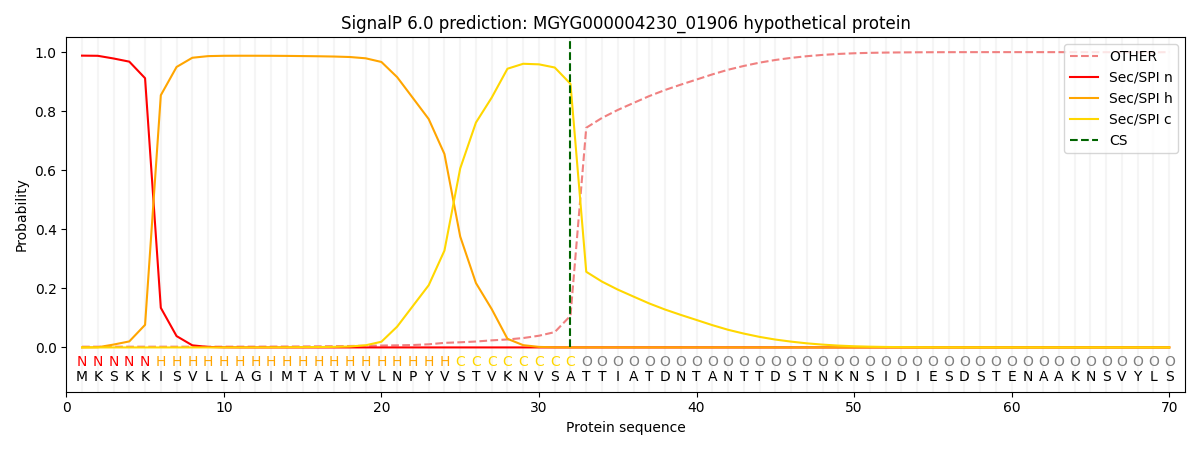
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004378 | 0.984316 | 0.010442 | 0.000365 | 0.000262 | 0.000211 |