You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004244_01731
You are here: Home > Sequence: MGYG000004244_01731
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
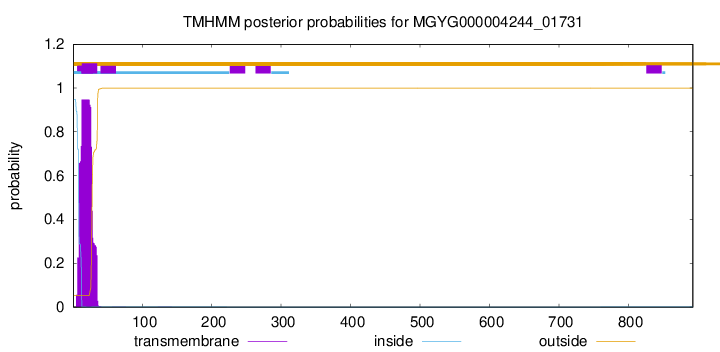
TMHMM annotations
Basic Information help
| Species | UMGS1601 sp900553335 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; UMGS1601; UMGS1601 sp900553335 | |||||||||||
| CAZyme ID | MGYG000004244_01731 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8917; End: 11598 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 57 | 299 | 1.1e-89 | 0.9915611814345991 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.63e-72 | 54 | 306 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| cd14852 | LD-carboxypeptidase | 1.10e-68 | 735 | 879 | 1 | 162 | L,D-carboxypeptidase DacB and LdcB, and related proteins. This L,D-carboxypeptidase family includes LdcB LD-Carboxypeptidase from Streptococcus pneumoniae, Bacillus anthracis, and Bacillus subtilis, and L,D-carboxypeptidase DacB from Streptococcus pneumonia and Lactococcus lactis. These enzymes are active against cell-wall-derived tetrapeptides and synthetic tetrapeptides lacking the sugar moiety but are inactive against tetrapeptides terminating in L-alanine. L,D-carboxypeptidase DacB plays a key role in the remodeling of S. pneumoniae peptidoglycan during cell division. It adopts a zinc-dependent carboxypeptidase fold and acts as an L,D-carboxypeptidase towards the tetrapeptide L-Ala-D-iGln-L-Lys-D-Ala of the peptidoglycan stem. This family also includes vanY D-Ala-D-Ala carboxypeptidase which is vancomycin-inducible and penicillin-resistant. VanY hydrolyzes depsipeptide- and D-alanyl-D-alanine-containing peptidoglycan precursors; it is insensitive to beta-lactams. All these enzymes belong to the MEROPS family M15 subfamily B. |
| pfam02557 | VanY | 2.26e-53 | 752 | 873 | 4 | 131 | D-alanyl-D-alanine carboxypeptidase. |
| COG1876 | LdcB | 5.13e-45 | 747 | 887 | 95 | 241 | LD-carboxypeptidase LdcB, LAS superfamily [Cell wall/membrane/envelope biogenesis]. |
| cd14849 | DD-dipeptidase_VanXYc | 1.03e-28 | 778 | 872 | 26 | 127 | D-Ala-D-Ala dipeptidase/D-Ala-D-Ala carboxypeptidase (VanXYc) and related proteins. VanXYc peptidase (also known as vanXY(C) peptidase, D-alanyl-D-alanine carboxypeptidase D,D-dipeptidase/D,D-carboxypeptidase, vancomycin resistance D,D-dipeptidase) is a Zn2+-dependent enzyme that mediates resistance to the antibiotic vancomycin in Enterococci. Some of the vancomycin resistance operons encode VanXY D,D-carboxypeptidase which hydrolyzes both, dipeptide (D-Ala-D-Ala) or pentapeptide (UDP-MurNac-L-Ala-D-Glu-L-Lys-D-Ala-D-Ala). It is a bifunctional enzyme that catalyzes D,D-peptidase and D,D-carboxypeptidase activities. VanXY has higher sequence similarity to VanY than with VanX and hydrolyzes D,D-dipeptides such as D-Ala-D-Ala, whereas VanY is inactive against this substrate; thus having a less restrictive active site to accommodate larger substrates such as UDP-MurNAc-pentapeptide[Ala]. This family belongs to the MEROPS family M15, subfamily B, and includes the D,D-dipeptidases VanXYg and VanXYe. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCO03766.1 | 1.01e-115 | 31 | 338 | 230 | 540 |
| ABV08875.1 | 1.22e-92 | 31 | 332 | 36 | 332 |
| AUO07708.1 | 3.38e-92 | 32 | 336 | 37 | 336 |
| AWW27905.1 | 7.16e-92 | 38 | 310 | 220 | 493 |
| AUS28000.1 | 1.15e-91 | 31 | 336 | 15 | 315 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 7.07e-98 | 38 | 336 | 5 | 297 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 3PZT_A | 1.32e-85 | 31 | 336 | 22 | 321 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
| 4XZB_A | 4.59e-85 | 38 | 336 | 4 | 300 | endo-glucanaseGsCelA P1 [Geobacillus sp. 70PC53] |
| 5WH8_A | 1.19e-84 | 34 | 336 | 2 | 304 | CellulaseCel5C_n [uncultured organism] |
| 4XZW_A | 6.37e-84 | 38 | 336 | 4 | 299 | Endo-glucanasechimera C10 [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10475 | 1.18e-82 | 31 | 336 | 27 | 326 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P07983 | 4.40e-82 | 31 | 336 | 27 | 326 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| P06565 | 3.51e-81 | 35 | 348 | 25 | 335 | Endoglucanase B OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=celB PE=3 SV=1 |
| O85465 | 5.28e-80 | 31 | 317 | 23 | 305 | Endoglucanase 5A OS=Salipaludibacillus agaradhaerens OX=76935 GN=cel5A PE=1 SV=1 |
| P23549 | 8.55e-80 | 31 | 336 | 27 | 326 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
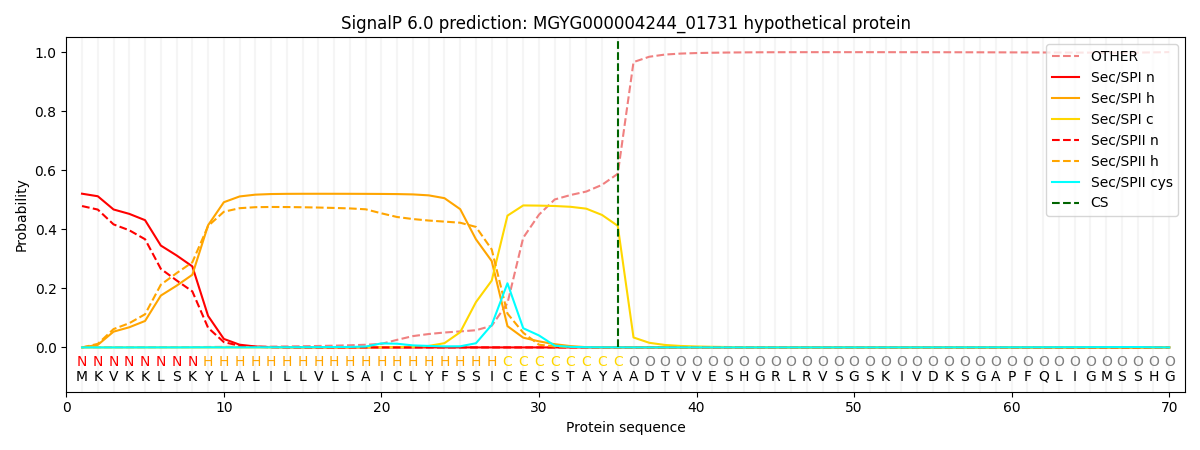
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001648 | 0.511497 | 0.486214 | 0.000231 | 0.000195 | 0.000199 |