You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004255_00406
You are here: Home > Sequence: MGYG000004255_00406
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Amulumruptor sp900547825 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Amulumruptor; Amulumruptor sp900547825 | |||||||||||
| CAZyme ID | MGYG000004255_00406 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | 1,4-alpha-glucan branching enzyme GlgB | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35151; End: 37892 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH13 | 412 | 714 | 1.1e-45 | 0.9431438127090301 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd11350 | AmyAc_4 | 3.27e-129 | 379 | 760 | 1 | 390 | Alpha amylase catalytic domain found in an uncharacterized protein family. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost the catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. |
| cd11325 | AmyAc_GTHase | 1.05e-49 | 373 | 710 | 12 | 362 | Alpha amylase catalytic domain found in Glycosyltrehalose trehalohydrolase (also called Maltooligosyl trehalose Trehalohydrolase). Glycosyltrehalose trehalohydrolase (GTHase) was discovered as part of a coupled system for the production of trehalose from soluble starch. In the first half of the reaction, glycosyltrehalose synthase (GTSase), an intramolecular glycosyl transferase, converts the glycosidic bond between the last two glucose residues of amylose from an alpha-1,4 bond to an alpha-1,1 bond, making a non-reducing glycosyl trehaloside. In the second half of the reaction, GTHase cleaves the alpha-1,4 glycosidic bond adjacent to the trehalose moiety to release trehalose and malto-oligosaccharide. Like isoamylase and other glycosidases that recognize branched oligosaccharides, GTHase contains an N-terminal extension and does not have the conserved calcium ion present in other alpha amylase family enzymes. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost the catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. Glycosyltrehalose Trehalohydrolase Maltooligosyltrehalose Trehalohydrolase |
| COG0296 | GlgB | 1.29e-44 | 255 | 709 | 25 | 472 | 1,4-alpha-glucan branching enzyme [Carbohydrate transport and metabolism]. |
| cd00551 | AmyAc_family | 7.51e-35 | 395 | 709 | 1 | 259 | Alpha amylase catalytic domain family. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; and C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost this catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. |
| cd11326 | AmyAc_Glg_debranch | 2.87e-32 | 379 | 553 | 1 | 205 | Alpha amylase catalytic domain found in glycogen debranching enzymes. Debranching enzymes facilitate the breakdown of glycogen through glucosyltransferase and glucosidase activity. These activities are performed by a single enzyme in mammals, yeast, and some bacteria, but by two distinct enzymes in Escherichia coli and other bacteria. Debranching enzymes perform two activities: 4-alpha-D-glucanotransferase (EC 2.4.1.25) and amylo-1,6-glucosidase (EC 3.2.1.33). 4-alpha-D-glucanotransferase catalyzes the endohydrolysis of 1,6-alpha-D-glucoside linkages at points of branching in chains of 1,4-linked alpha-D-glucose residues. Amylo-alpha-1,6-glucosidase catalyzes the endohydrolysis of 1,6-alpha-D-glucoside linkages at points of branching in chains of 1,4-linked alpha-D-glucose residues. In Escherichia coli, GlgX is the debranching enzyme and malQ is the 4-alpha-glucanotransferase. TreX, an archaeal glycogen-debranching enzyme has dual activities like mammals and yeast, but is structurally similar to GlgX. TreX exists in two oligomeric states, a dimer and tetramer. Isoamylase (EC 3.2.1.68) is one of the starch-debranching enzymes that catalyzes the hydrolysis of alpha-1,6-glucosidic linkages specific in alpha-glucans such as amylopectin or glycogen and their beta-limit dextrins. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost the catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD35300.1 | 5.01e-256 | 1 | 913 | 1 | 927 |
| QIM10833.1 | 1.11e-252 | 24 | 913 | 11 | 908 |
| QCD42371.1 | 1.09e-200 | 256 | 912 | 1 | 683 |
| QQR08212.1 | 3.10e-191 | 20 | 818 | 26 | 758 |
| ANU64421.1 | 3.10e-191 | 20 | 818 | 26 | 758 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3M07_A | 9.48e-27 | 379 | 562 | 122 | 295 | 1.4Angstrom Resolution Crystal Structure of Putative alpha Amylase from Salmonella typhimurium. [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 1EHA_A | 7.85e-23 | 263 | 559 | 8 | 259 | CRYSTALSTRUCTURE OF GLYCOSYLTREHALOSE TREHALOHYDROLASE FROM SULFOLOBUS SOLFATARICUS [Saccharolobus solfataricus] |
| 3VGG_A | 7.85e-23 | 263 | 559 | 8 | 259 | Crystalstructure of glycosyltrehalose trehalohydrolase (E283Q) complexed with maltoheptaose [Saccharolobus solfataricus],3VGH_A Crystal structure of glycosyltrehalose trehalohydrolase (E283Q) complexed with maltotriosyltrehalose [Saccharolobus solfataricus] |
| 1EH9_A | 7.85e-23 | 263 | 559 | 8 | 259 | CrystalStructure Of Sulfolobus Solfataricus Glycosyltrehalose Trehalohydrolase [Saccharolobus solfataricus],3VGB_A Crystal structure of glycosyltrehalose trehalohydrolase (GTHase) from Sulfolobus solfataricus KM1 [Saccharolobus solfataricus] |
| 3VGD_A | 2.44e-22 | 263 | 559 | 8 | 259 | Ctystalstructure of glycosyltrehalose trehalohydrolase (D252E) [Saccharolobus solfataricus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B9G434 | 1.81e-23 | 326 | 557 | 195 | 450 | Isoamylase 3, chloroplastic OS=Oryza sativa subsp. japonica OX=39947 GN=ISA3 PE=2 SV=1 |
| P9WQ23 | 2.15e-23 | 384 | 574 | 86 | 269 | Malto-oligosyltrehalose trehalohydrolase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=treZ PE=1 SV=1 |
| P9WQ22 | 2.15e-23 | 384 | 574 | 86 | 269 | Malto-oligosyltrehalose trehalohydrolase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=treZ PE=3 SV=1 |
| Q9AJN6 | 6.49e-23 | 379 | 559 | 85 | 257 | Malto-oligosyltrehalose trehalohydrolase OS=Arthrobacter ramosus OX=1672 GN=treZ PE=3 SV=1 |
| Q9M0S5 | 2.12e-22 | 369 | 557 | 206 | 433 | Isoamylase 3, chloroplastic OS=Arabidopsis thaliana OX=3702 GN=ISA3 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
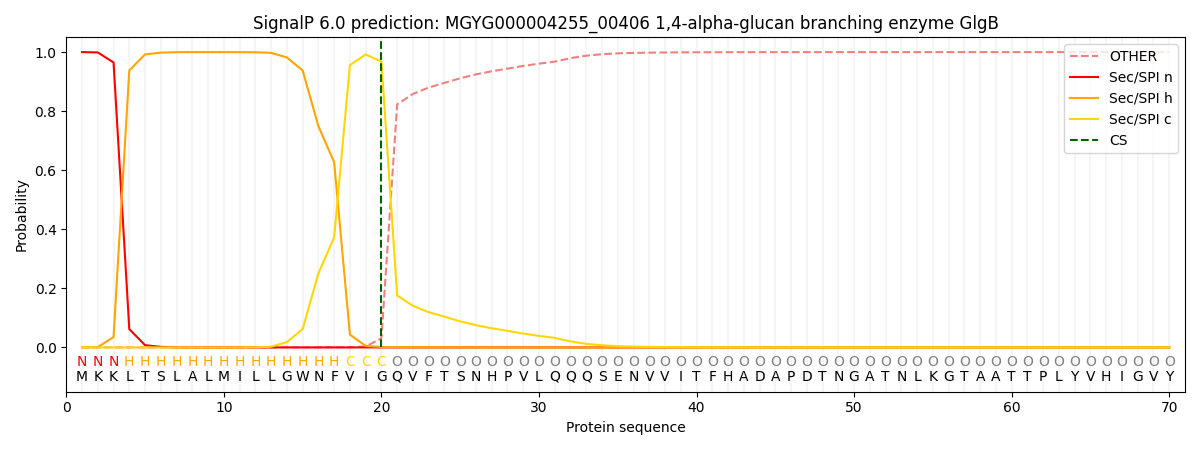
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000827 | 0.998100 | 0.000303 | 0.000260 | 0.000252 | 0.000239 |
