You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004279_00201
You are here: Home > Sequence: MGYG000004279_00201
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
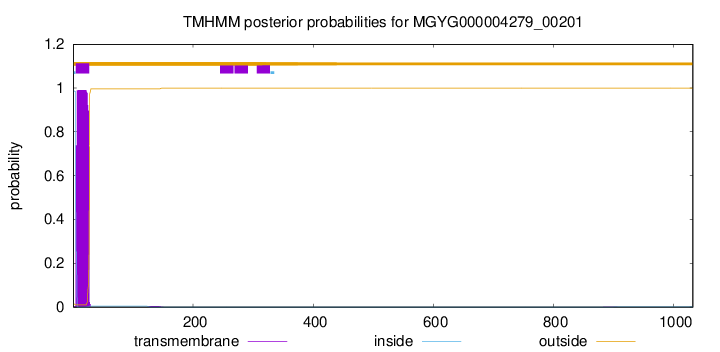
TMHMM annotations
Basic Information help
| Species | UMGS1484 sp900552285 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; UMGS1484; UMGS1484 sp900552285 | |||||||||||
| CAZyme ID | MGYG000004279_00201 | |||||||||||
| CAZy Family | GH36 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17124; End: 20222 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH36 | 720 | 1015 | 6.7e-19 | 0.40988372093023256 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14256 | Dockerin_I | 1.29e-09 | 273 | 326 | 1 | 54 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam16874 | Glyco_hydro_36C | 7.32e-09 | 946 | 1029 | 1 | 78 | Glycosyl hydrolase family 36 C-terminal domain. This domain is found at the C-terminus of many family 36 glycoside hydrolases. It has a beta-sandwich structure with a Greek key motif. |
| cd14791 | GH36 | 1.24e-08 | 632 | 860 | 32 | 244 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| COG3345 | GalA | 6.44e-07 | 632 | 1019 | 322 | 670 | Alpha-galactosidase [Carbohydrate transport and metabolism]. |
| pfam02065 | Melibiase | 1.09e-05 | 632 | 860 | 71 | 285 | Melibiase. Glycoside hydrolase families GH27, GH31 and GH36 form the glycoside hydrolase clan GH-D. Glycoside hydrolase family 36 can be split into 11 families, GH36A to GH36K. This family includes enzymes from GH36A-B and GH36D-K and from GH27. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGQ94273.1 | 1.58e-129 | 344 | 1030 | 346 | 1029 |
| AEE97820.1 | 1.37e-17 | 570 | 1026 | 248 | 684 |
| CED94212.1 | 1.36e-14 | 632 | 1031 | 328 | 695 |
| AYQ72355.1 | 1.05e-12 | 455 | 1031 | 68 | 653 |
| BBH24245.1 | 2.02e-12 | 40 | 186 | 1097 | 1246 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6JHP_A | 1.86e-09 | 720 | 1031 | 468 | 757 | Crystalstructure of the glycoside hydrolase family 36 alpha-galactosidase from Paecilomyces thermophila [Paecilomyces sp. 'thermophila'],6JHP_B Crystal structure of the glycoside hydrolase family 36 alpha-galactosidase from Paecilomyces thermophila [Paecilomyces sp. 'thermophila'],6JHP_C Crystal structure of the glycoside hydrolase family 36 alpha-galactosidase from Paecilomyces thermophila [Paecilomyces sp. 'thermophila'],6JHP_D Crystal structure of the glycoside hydrolase family 36 alpha-galactosidase from Paecilomyces thermophila [Paecilomyces sp. 'thermophila'] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5AU92 | 1.01e-08 | 599 | 1030 | 346 | 748 | Alpha-galactosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=aglC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
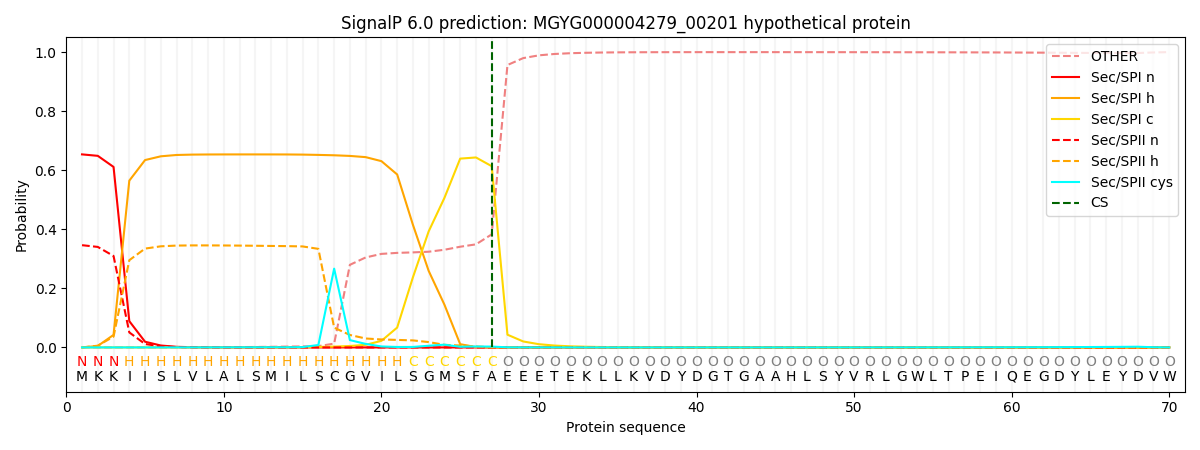
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001103 | 0.640946 | 0.357014 | 0.000358 | 0.000294 | 0.000258 |