You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004281_00835
You are here: Home > Sequence: MGYG000004281_00835
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp900552465 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp900552465 | |||||||||||
| CAZyme ID | MGYG000004281_00835 | |||||||||||
| CAZy Family | GH31 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 43159; End: 43851 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14752 | GH31_N | 3.67e-22 | 85 | 221 | 8 | 122 | N-terminal domain of glycosyl hydrolase family 31 (GH31). This family is found N-terminal to the glycosyl-hydrolase domain of Glycoside hydrolase family 31 (GH31). GH31 includes the glycoside hydrolases alpha-glucosidase (EC 3.2.1.20), alpha-1,3-glucosidase (EC 3.2.1.84), alpha-xylosidase (EC 3.2.1.177), sucrase-isomaltase (EC 3.2.1.48 and EC 3.2.1.10), as well as alpha-glucan lyase (EC 4.2.2.13). All GH31 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. In most cases, the pyranose moiety recognized in subsite-1 of the substrate binding site is an alpha-D-glucose, though some GH31 family members show a preference for alpha-D-xylose. Several GH31 enzymes can accommodate both glucose and xylose and different levels of discrimination between the two have been observed. Most characterized GH31 enzymes are alpha-glucosidases. In mammals, GH31 members with alpha-glucosidase activity are implicated in at least three distinct biological processes. The lysosomal acid alpha-glucosidase (GAA) is essential for glycogen degradation and a deficiency or malfunction of this enzyme causes glycogen storage disease II, also known as Pompe disease. In the endoplasmic reticulum, alpha-glucosidase II catalyzes the second step in the N-linked oligosaccharide processing pathway that constitutes part of the quality control system for glycoprotein folding and maturation. The intestinal enzymes sucrase-isomaltase (SI) and maltase-glucoamylase (MGAM) play key roles in the final stage of carbohydrate digestion, making alpha-glucosidase inhibitors useful in the treatment of type 2 diabetes. GH31 alpha-glycosidases are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues of the catalytic domain have been identified as the catalytic nucleophile and the acid/base, respectively. A loop of the N-terminal beta-sandwich domain is part of the active site pocket. |
| PRK10658 | PRK10658 | 6.60e-08 | 84 | 229 | 144 | 266 | putative alpha-glucosidase; Provisional |
| COG1501 | YicI | 5.32e-07 | 84 | 229 | 140 | 264 | Alpha-glucosidase, glycosyl hydrolase family GH31 [Carbohydrate transport and metabolism]. |
| PRK10426 | PRK10426 | 5.63e-06 | 74 | 153 | 59 | 157 | alpha-glucosidase; Provisional |
| pfam13802 | Gal_mutarotas_2 | 1.27e-05 | 96 | 151 | 1 | 61 | Galactose mutarotase-like. This family is found N-terminal to glycosyl-hydrolase domains, and appears to be similar to the galactose mutarotase superfamily. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ59521.1 | 6.97e-108 | 4 | 230 | 6 | 230 |
| QUT89436.1 | 6.97e-108 | 4 | 230 | 6 | 230 |
| ALJ60418.1 | 4.17e-99 | 35 | 230 | 30 | 225 |
| AEI50598.1 | 1.16e-95 | 11 | 230 | 9 | 228 |
| AXE16679.1 | 3.38e-94 | 7 | 230 | 6 | 228 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4XPO_A | 1.74e-83 | 30 | 230 | 31 | 231 | Crystalstructure of a novel alpha-galactosidase from Pedobacter saltans [Pseudopedobacter saltans],4XPP_A Crystal structure of Pedobacter saltans GH31 alpha-galactosidase complexed with D-galactose [Pseudopedobacter saltans],4XPQ_A Crystal structure of Pedobacter saltans GH31 alpha-galactosidase complexed with L-fucose [Pseudopedobacter saltans] |
| 4XPR_A | 1.74e-83 | 30 | 230 | 31 | 231 | Crystalstructure of the mutant D365A of Pedobacter saltans GH31 alpha-galactosidase [Pseudopedobacter saltans],4XPS_A Crystal structure of the mutant D365A of Pedobacter saltans GH31 alpha-galactosidase complexed with p-nitrophenyl-alpha-galactopyranoside [Pseudopedobacter saltans] |
| 5I0D_A | 4.18e-10 | 84 | 229 | 230 | 363 | Cycloalternan-formingenzyme from Listeria monocytogenes in complex with cycloalternan [Listeria monocytogenes EGD-e],5I0D_B Cycloalternan-forming enzyme from Listeria monocytogenes in complex with cycloalternan [Listeria monocytogenes EGD-e] |
| 4KMQ_A | 4.19e-10 | 84 | 229 | 251 | 384 | 1.9Angstrom resolution crystal structure of uncharacterized protein lmo2446 from Listeria monocytogenes EGD-e [Listeria monocytogenes EGD-e],4KWU_A 1.9 Angstrom resolution crystal structure of uncharacterized protein lmo2446 from Listeria monocytogenes EGD-e in complex with alpha-D-glucose, beta-D-glucose, magnesium and calcium [Listeria monocytogenes EGD-e],5HPO_A Cycloalternan-forming enzyme from Listeria monocytogenes in complex with maltopentaose [Listeria monocytogenes EGD-e],5HXM_A Cycloalternan-forming enzyme from Listeria monocytogenes in complex with panose [Listeria monocytogenes] |
| 5F7U_A | 7.59e-10 | 84 | 229 | 230 | 363 | Cycloalternan-formingenzyme from Listeria monocytogenes in complex with pentasaccharide substrate [Listeria monocytogenes EGD-e] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P96793 | 1.03e-07 | 96 | 229 | 157 | 265 | Alpha-xylosidase XylQ OS=Lactiplantibacillus pentosus OX=1589 GN=xylQ PE=1 SV=1 |
| Q5AW25 | 8.75e-06 | 67 | 229 | 152 | 285 | Alpha-xylosidase OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=agdD PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
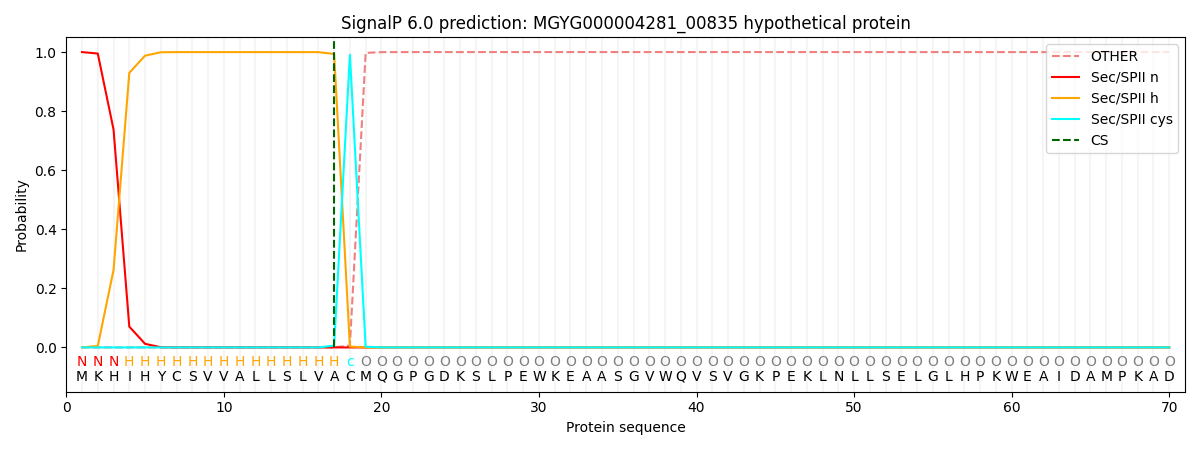
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000008 | 1.000039 | 0.000000 | 0.000000 | 0.000000 |
