You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004287_01777
You are here: Home > Sequence: MGYG000004287_01777
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
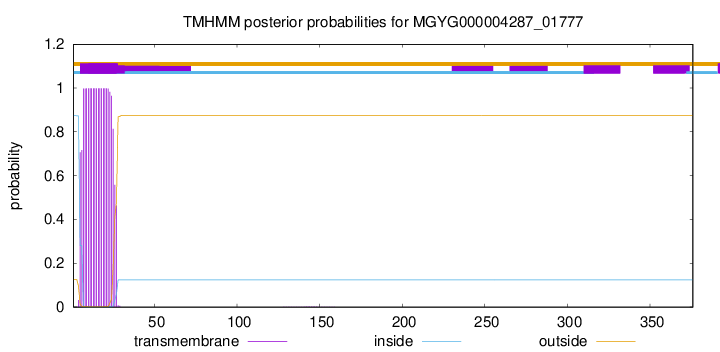
TMHMM annotations
Basic Information help
| Species | Limosilactobacillus caviae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Limosilactobacillus; Limosilactobacillus caviae | |||||||||||
| CAZyme ID | MGYG000004287_01777 | |||||||||||
| CAZy Family | GH8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 28869; End: 29999 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01270 | Glyco_hydro_8 | 3.90e-13 | 36 | 328 | 6 | 277 | Glycosyl hydrolases family 8. |
| COG3405 | BcsZ | 2.37e-12 | 18 | 374 | 8 | 344 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
| PRK11097 | PRK11097 | 0.006 | 69 | 296 | 54 | 270 | cellulase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAG25983.1 | 5.59e-209 | 1 | 376 | 1 | 379 |
| QPB66875.1 | 5.59e-209 | 1 | 376 | 1 | 379 |
| AKP01773.1 | 5.59e-209 | 1 | 376 | 1 | 379 |
| QLQ62144.1 | 5.59e-209 | 1 | 376 | 1 | 379 |
| ABQ83804.1 | 5.59e-209 | 1 | 376 | 1 | 379 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6VC5_A | 8.24e-07 | 69 | 282 | 33 | 231 | 1.6Angstrom Resolution Crystal Structure of endoglucanase from Komagataeibacter sucrofermentans [Komagataeibacter sucrofermentans] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
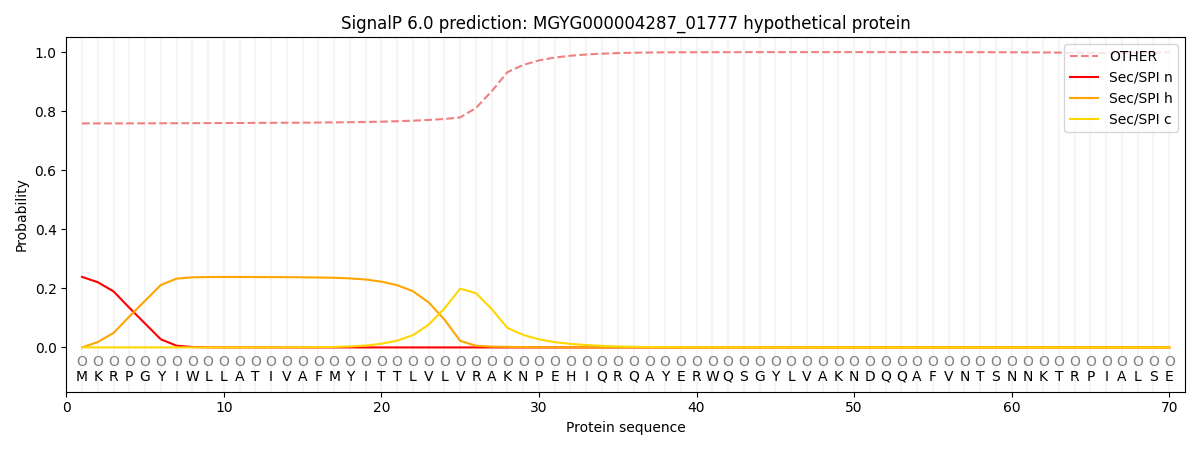
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.770913 | 0.223964 | 0.002201 | 0.000545 | 0.000343 | 0.002051 |