You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004303_00528
You are here: Home > Sequence: MGYG000004303_00528
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
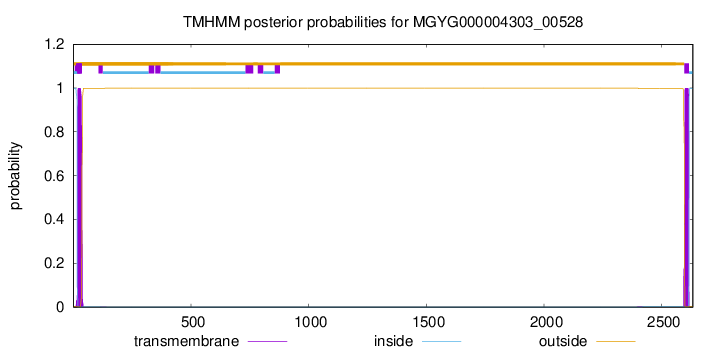
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RF39; UBA660; RUG13868; | |||||||||||
| CAZyme ID | MGYG000004303_00528 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 67169; End: 75067 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 59 | 203 | 4.8e-24 | 0.9850746268656716 |
| CBM32 | 912 | 1030 | 9.9e-17 | 0.8306451612903226 |
| CBM32 | 1229 | 1352 | 3.3e-16 | 0.7903225806451613 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 2.51e-23 | 56 | 203 | 1 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 1.37e-13 | 58 | 203 | 5 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam00754 | F5_F8_type_C | 9.83e-10 | 913 | 1026 | 3 | 110 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 5.95e-09 | 1236 | 1346 | 14 | 115 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 4.84e-04 | 1236 | 1344 | 25 | 129 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMN35280.1 | 2.05e-195 | 55 | 1045 | 46 | 988 |
| AQW23377.1 | 3.89e-195 | 55 | 1045 | 46 | 988 |
| ATD49073.1 | 4.51e-195 | 55 | 1045 | 51 | 993 |
| BCL58565.1 | 1.44e-132 | 51 | 1047 | 74 | 1051 |
| SQG16429.1 | 2.73e-108 | 51 | 1038 | 134 | 1085 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JS4_A | 2.16e-11 | 52 | 203 | 606 | 752 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
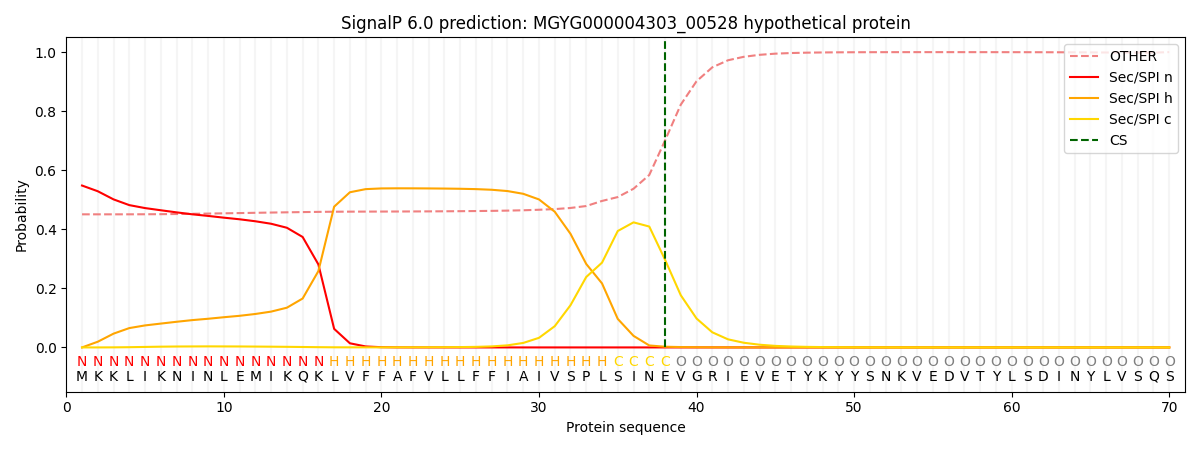
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.465809 | 0.530610 | 0.001796 | 0.000284 | 0.000325 | 0.001176 |