You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004314_00203
You are here: Home > Sequence: MGYG000004314_00203
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
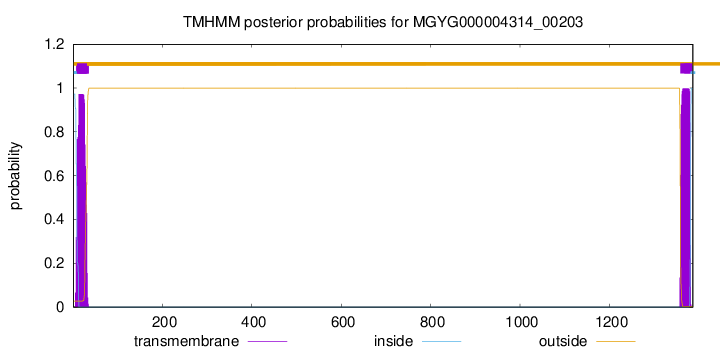
TMHMM annotations
Basic Information help
| Species | Varibaculum timonense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Varibaculum; Varibaculum timonense | |||||||||||
| CAZyme ID | MGYG000004314_00203 | |||||||||||
| CAZy Family | GH29 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1158; End: 5321 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH29 | 491 | 859 | 4.8e-61 | 0.884393063583815 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3669 | AfuC | 5.72e-50 | 501 | 995 | 11 | 430 | Alpha-L-fucosidase [Carbohydrate transport and metabolism]. |
| smart00812 | Alpha_L_fucos | 3.30e-31 | 527 | 858 | 61 | 325 | Alpha-L-fucosidase. O-Glycosyl hydrolases (EC 3.2.1.-) are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A classification system for glycosyl hydrolases, based on sequence similarity, has led to the definition of 85 different families. This classification is available on the CAZy (CArbohydrate-Active EnZymes) web site. Because the fold of proteins is better conserved than their sequences, some of the families can be grouped in 'clans'. Family 29 encompasses alpha-L-fucosidases, which is a lysosomal enzyme responsible for hydrolyzing the alpha-1,6-linked fucose joined to the reducing-end N-acetylglucosamine of the carbohydrate moieties of glycoproteins. Deficiency of alpha-L-fucosidase results in the lysosomal storage disease fucosidosis. |
| pfam01120 | Alpha_L_fucos | 1.58e-28 | 540 | 719 | 80 | 226 | Alpha-L-fucosidase. |
| pfam00754 | F5_F8_type_C | 9.05e-15 | 1129 | 1267 | 2 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| NF033201 | Vip_LPXTG_Lm | 8.09e-05 | 1276 | 1383 | 281 | 387 | cell invasion LPXTG protein Vip. Vip (Virulence protein), like the LPXTG-type internalins, is an LPXTG-anchored surface protein of the mammalian cell-invading pathogen Listeria monocytogenes, but absent from the related species Listeria innocua. For certain cell types, Vip is required for Listeria's ability to invade. It appears to bind the endoplasmic reticulum (ER) resident chaperone Gp96 as its receptor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AXM91280.1 | 0.0 | 17 | 1289 | 25 | 1283 |
| BBA47373.1 | 0.0 | 17 | 1289 | 25 | 1283 |
| QRI58322.1 | 0.0 | 17 | 1289 | 25 | 1283 |
| BBA55614.1 | 0.0 | 17 | 1289 | 25 | 1283 |
| AFL04933.1 | 0.0 | 17 | 1289 | 25 | 1283 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ZRX_A | 2.00e-69 | 499 | 1028 | 30 | 489 | Crystalstructure of a putative alpha-L-fucosidase (BACOVA_04357) from Bacteroides ovatus ATCC 8483 at 1.59 A resolution [Bacteroides ovatus ATCC 8483] |
| 3UES_A | 1.93e-68 | 496 | 991 | 14 | 472 | Crystalstructure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis complexed with deoxyfuconojirimycin [Bifidobacterium longum subsp. infantis ATCC 15697 = JCM 1222 = DSM 20088],3UES_B Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis complexed with deoxyfuconojirimycin [Bifidobacterium longum subsp. infantis ATCC 15697 = JCM 1222 = DSM 20088] |
| 3MO4_A | 2.34e-67 | 496 | 991 | 16 | 474 | Thecrystal structure of an alpha-(1-3,4)-fucosidase from Bifidobacterium longum subsp. infantis ATCC 15697 [Bifidobacterium longum subsp. infantis ATCC 15697 = JCM 1222 = DSM 20088],3MO4_B The crystal structure of an alpha-(1-3,4)-fucosidase from Bifidobacterium longum subsp. infantis ATCC 15697 [Bifidobacterium longum subsp. infantis ATCC 15697 = JCM 1222 = DSM 20088] |
| 3UET_A | 1.39e-66 | 496 | 991 | 14 | 472 | Crystalstructure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis D172A/E217A mutant complexed with lacto-N-fucopentaose II [Bifidobacterium longum subsp. infantis ATCC 15697 = JCM 1222 = DSM 20088],3UET_B Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis D172A/E217A mutant complexed with lacto-N-fucopentaose II [Bifidobacterium longum subsp. infantis ATCC 15697 = JCM 1222 = DSM 20088] |
| 6ORG_A | 1.01e-52 | 501 | 995 | 11 | 448 | Crystalstructure of SpGH29 [Streptococcus pneumoniae TIGR4],6ORG_B Crystal structure of SpGH29 [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8GW72 | 1.50e-58 | 501 | 995 | 38 | 477 | Alpha-L-fucosidase 1 OS=Arabidopsis thaliana OX=3702 GN=FUC1 PE=1 SV=2 |
| Q7XUR3 | 2.47e-44 | 516 | 985 | 55 | 466 | Putative alpha-L-fucosidase 1 OS=Oryza sativa subsp. japonica OX=39947 GN=Os04g0560400 PE=3 SV=2 |
| Q02834 | 1.03e-08 | 1123 | 1270 | 506 | 644 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| Q99LJ1 | 2.22e-08 | 516 | 732 | 47 | 279 | Tissue alpha-L-fucosidase OS=Mus musculus OX=10090 GN=Fuca1 PE=1 SV=1 |
| P0DTR4 | 1.00e-07 | 1037 | 1269 | 419 | 643 | A type blood N-acetyl-alpha-D-galactosamine deacetylase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
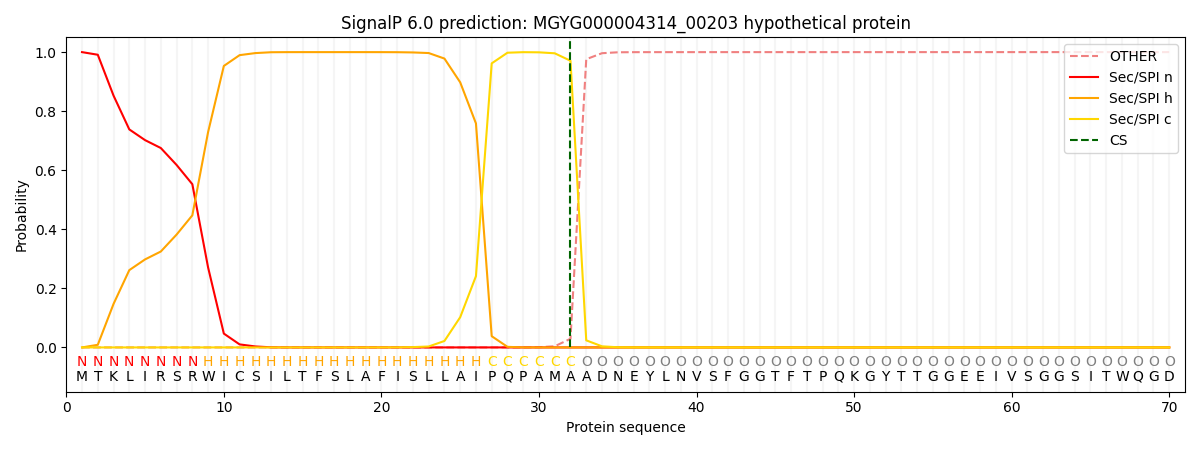
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000465 | 0.998566 | 0.000242 | 0.000249 | 0.000212 | 0.000204 |