You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004365_00467
You are here: Home > Sequence: MGYG000004365_00467
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; UBA7173; | |||||||||||
| CAZyme ID | MGYG000004365_00467 | |||||||||||
| CAZy Family | GH63 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17056; End: 18966 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH63 | 275 | 623 | 1.1e-27 | 0.356140350877193 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK10137 | PRK10137 | 5.83e-17 | 273 | 606 | 342 | 765 | alpha-glucosidase; Provisional |
| pfam01204 | Trehalase | 7.31e-16 | 281 | 604 | 131 | 478 | Trehalase. Trehalase (EC:3.2.1.28) is known to recycle trehalose to glucose. Trehalose is a physiological hallmark of heat-shock response in yeast and protects of proteins and membranes against a variety of stresses. This family is found in conjunction with pfam07492 in fungi. |
| pfam03200 | Glyco_hydro_63 | 1.10e-10 | 499 | 626 | 352 | 492 | Glycosyl hydrolase family 63 C-terminal domain. This is a family of eukaryotic enzymes belonging to glycosyl hydrolase family 63. They catalyze the specific cleavage of the non-reducing terminal glucose residue from Glc(3)Man(9)GlcNAc(2). Mannosyl oligosaccharide glucosidase EC:3.2.1.106 is the first enzyme in the N-linked oligosaccharide processing pathway. This family represents the C-terminal catalytic domain. |
| COG1626 | TreA | 2.24e-10 | 278 | 600 | 172 | 510 | Neutral trehalase [Carbohydrate transport and metabolism]. |
| COG3408 | GDB1 | 5.90e-07 | 264 | 579 | 269 | 564 | Glycogen debranching enzyme (alpha-1,6-glucosidase) [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDO68990.1 | 9.94e-274 | 1 | 634 | 1 | 633 |
| QUT46474.1 | 1.53e-270 | 1 | 633 | 1 | 632 |
| QRQ47302.1 | 4.37e-270 | 1 | 633 | 1 | 632 |
| ANH81205.1 | 3.00e-231 | 9 | 629 | 5 | 632 |
| QNK58860.1 | 7.87e-172 | 30 | 629 | 16 | 614 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3W7S_A | 2.13e-12 | 273 | 625 | 315 | 751 | Escherichiacoli K12 YgjK complexed with glucose [Escherichia coli K-12],3W7S_B Escherichia coli K12 YgjK complexed with glucose [Escherichia coli K-12],3W7T_A Escherichia coli K12 YgjK complexed with mannose [Escherichia coli K-12],3W7T_B Escherichia coli K12 YgjK complexed with mannose [Escherichia coli K-12],3W7U_A Escherichia coli K12 YgjK complexed with galactose [Escherichia coli K-12],3W7U_B Escherichia coli K12 YgjK complexed with galactose [Escherichia coli K-12] |
| 3W7X_A | 8.45e-12 | 273 | 625 | 315 | 751 | Crystalstructure of E. coli YgjK D324N complexed with melibiose [Escherichia coli K-12],3W7X_B Crystal structure of E. coli YgjK D324N complexed with melibiose [Escherichia coli K-12],5CA3_A Crystal structure of the glycosynthase mutant D324N of Escherichia coli GH63 glycosidase in complex with glucose and lactose [Escherichia coli K-12],5CA3_B Crystal structure of the glycosynthase mutant D324N of Escherichia coli GH63 glycosidase in complex with glucose and lactose [Escherichia coli K-12] |
| 3W7W_A | 1.11e-11 | 273 | 625 | 315 | 751 | Crystalstructure of E. coli YgjK E727A complexed with 2-O-alpha-D-glucopyranosyl-alpha-D-galactopyranose [Escherichia coli K-12],3W7W_B Crystal structure of E. coli YgjK E727A complexed with 2-O-alpha-D-glucopyranosyl-alpha-D-galactopyranose [Escherichia coli K-12],5GW7_A Crystal structure of the glycosynthase mutant E727A of Escherichia coli GH63 glycosidase in complex with glucose and lactose [Escherichia coli K-12],5GW7_B Crystal structure of the glycosynthase mutant E727A of Escherichia coli GH63 glycosidase in complex with glucose and lactose [Escherichia coli K-12] |
| 3D3I_A | 5.84e-11 | 273 | 624 | 316 | 751 | Crystalstructural of Escherichia coli K12 YgjK, a glucosidase belonging to glycoside hydrolase family 63 [Escherichia coli K-12],3D3I_B Crystal structural of Escherichia coli K12 YgjK, a glucosidase belonging to glycoside hydrolase family 63 [Escherichia coli K-12] |
| 6XUX_A | 5.71e-10 | 427 | 625 | 97 | 300 | ChainA, Nanobody,Glucosidase YgjK,Glucosidase YgjK,Nanobody [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P42592 | 1.18e-11 | 273 | 625 | 338 | 774 | Glucosidase YgjK OS=Escherichia coli (strain K12) OX=83333 GN=ygjK PE=1 SV=1 |
| Q757L1 | 2.17e-09 | 401 | 616 | 458 | 677 | Probable trehalase OS=Ashbya gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056) OX=284811 GN=NTH2 PE=3 SV=1 |
| P49381 | 8.68e-09 | 473 | 616 | 549 | 693 | Cytosolic neutral trehalase OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=NTH1 PE=1 SV=1 |
| O42783 | 1.14e-08 | 278 | 616 | 290 | 682 | Cytosolic neutral trehalase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=treB PE=2 SV=2 |
| O42777 | 7.84e-08 | 278 | 616 | 296 | 687 | Cytosolic neutral trehalase OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=treB PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
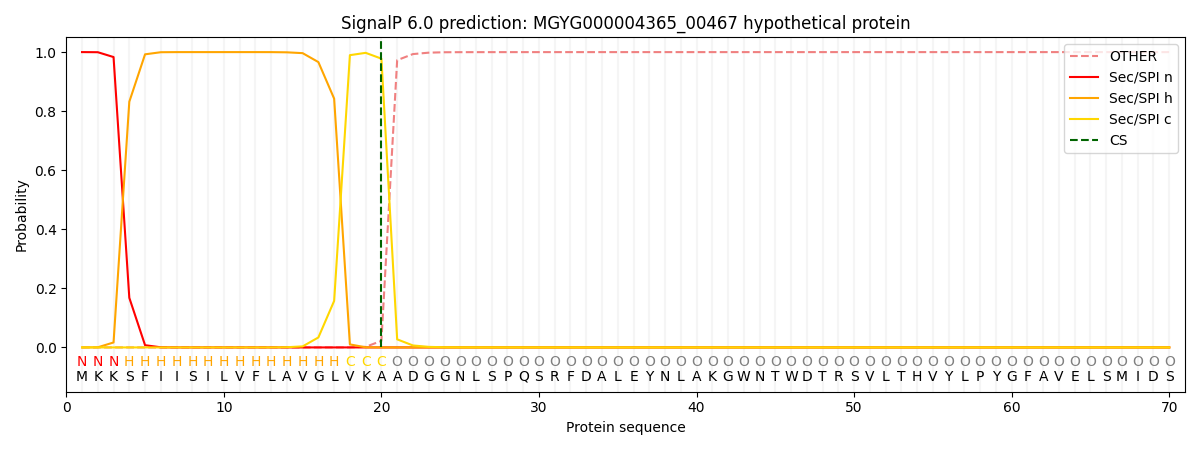
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000337 | 0.998801 | 0.000272 | 0.000185 | 0.000199 | 0.000182 |
